| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 15,571,817 – 15,571,911 |
| Length | 94 |
| Max. P | 0.653210 |

| Location | 15,571,817 – 15,571,911 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 83.61 |
| Mean single sequence MFE | -34.58 |
| Consensus MFE | -23.16 |
| Energy contribution | -24.97 |
| Covariance contribution | 1.81 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.653210 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
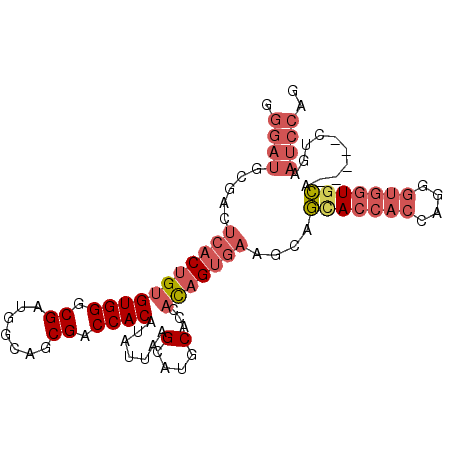
>2R_DroMel_CAF1 15571817 94 + 20766785 GGGAUGCGACUCACUGUGUGGGCGAUGGCAGCGACCACAUAUUAAGCAUGCACCACAGUGAGACAGAACCACCAGGGUGGUACA-----CUGAAUCCAG .((((....(((((((((.(.(((...((................)).))).)))))))))).(((.(((((....)))))...-----))).)))).. ( -32.29) >DroSec_CAF1 365277 94 + 1 GGGAUGCGACUCACUGUGUGGGCGAUGGCAGCGACCACAUAUUAAGCAUGCACCACAGAGAAGCAGCACCACCAUGGUGUUGCA-----CAGAAUCCAG .((((.(......((.(((((......(((((.............)).))).))))).))..((((((((.....)))))))).-----..).)))).. ( -32.92) >DroSim_CAF1 345210 94 + 1 GGGAUGCGACUCACUGUGUGGGCGAUGGCAGCGACCACAUAUUAAGCAUGCACCACAGUGAAGCAGCACCACCAGGGUGGUGCA-----CUGAAUCCAG .((((((...((((((((.(.(((...((................)).))).))))))))).)).(((((((....))))))).-----....)))).. ( -36.39) >DroEre_CAF1 353191 93 + 1 GGGAUGCGACUCACUGUGUGGGCGA-GGCAGCGACCACAUAUUAAGCAUGCACCACAGUGAAGCAGCACCACCAGGGUGGUGCA-----CUGCAUCCAG .(((((((..(((((((((((.((.-.....)).)))).......(....)...)))))))..).(((((((....))))))).-----..)))))).. ( -41.90) >DroYak_CAF1 368273 94 + 1 GGGAUGCGACUCACUGUGUGGGCGAAUGCAGCGACCACAUAUUAAGCAUGCACCACAGUGAAACAGCACCACCAGGGUGGUGCA-----CUGCAUCCAG .(((((((..((((((((.(.((..((((................)))))).)))))))))....(((((((....))))))).-----.))))))).. ( -41.59) >DroAna_CAF1 340337 83 + 1 --------ACUCACUGUGUGGGCGA--GCAGCGACCACAUAUUAAGAAGGCAUCUUCGU-----ACUACGAGUAGGG-GGUGGAAGAAUCUCUAUCAAG --------((((..(((((((.((.--....)).)))))))....((((....))))..-----.....))))....-((((((......))))))... ( -22.40) >consensus GGGAUGCGACUCACUGUGUGGGCGAUGGCAGCGACCACAUAUUAAGCAUGCACCACAGUGAAGCAGCACCACCAGGGUGGUGCA_____CUGAAUCCAG .((((.....(((((((((((.((.......)).)))).......(....)...)))))))....(((((((....)))))))..........)))).. (-23.16 = -24.97 + 1.81)

| Location | 15,571,817 – 15,571,911 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 83.61 |
| Mean single sequence MFE | -32.05 |
| Consensus MFE | -22.14 |
| Energy contribution | -23.87 |
| Covariance contribution | 1.72 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.69 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
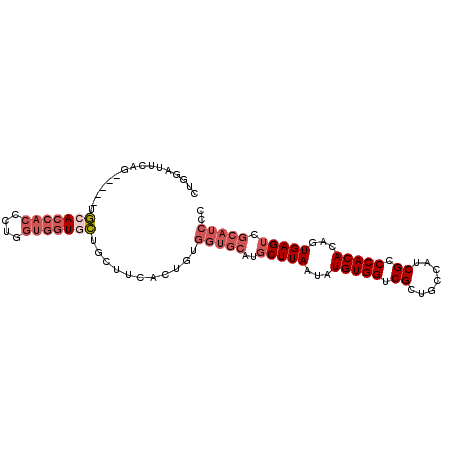
>2R_DroMel_CAF1 15571817 94 - 20766785 CUGGAUUCAG-----UGUACCACCCUGGUGGUUCUGUCUCACUGUGGUGCAUGCUUAAUAUGUGGUCGCUGCCAUCGCCCACACAGUGAGUCGCAUCCC ..((((.(((-----...(((((....))))).))(.((((((((((.((((.....))..(((((....))))).)).).))))))))).)).)))). ( -32.00) >DroSec_CAF1 365277 94 - 1 CUGGAUUCUG-----UGCAACACCAUGGUGGUGCUGCUUCUCUGUGGUGCAUGCUUAAUAUGUGGUCGCUGCCAUCGCCCACACAGUGAGUCGCAUCCC .(((((((..-----.(((.((((.....)))).)))..((.(((((.((((.....))..(((((....))))).))))))).)).))))).)).... ( -26.60) >DroSim_CAF1 345210 94 - 1 CUGGAUUCAG-----UGCACCACCCUGGUGGUGCUGCUUCACUGUGGUGCAUGCUUAAUAUGUGGUCGCUGCCAUCGCCCACACAGUGAGUCGCAUCCC ..((((.(((-----((((((((....))))))).)))(((((((((.((((.....))..(((((....))))).)).).))))))))...).)))). ( -35.00) >DroEre_CAF1 353191 93 - 1 CUGGAUGCAG-----UGCACCACCCUGGUGGUGCUGCUUCACUGUGGUGCAUGCUUAAUAUGUGGUCGCUGCC-UCGCCCACACAGUGAGUCGCAUCCC ..((((((((-----((((((((....))))))).)))((((((((..(((((.....)))))((.((.....-.)).)).))))))))...)))))). ( -41.80) >DroYak_CAF1 368273 94 - 1 CUGGAUGCAG-----UGCACCACCCUGGUGGUGCUGUUUCACUGUGGUGCAUGCUUAAUAUGUGGUCGCUGCAUUCGCCCACACAGUGAGUCGCAUCCC ..((((((..-----.(((((((....))))))).(.((((((((((((.((((................)))).))))...)))))))).))))))). ( -42.59) >DroAna_CAF1 340337 83 - 1 CUUGAUAGAGAUUCUUCCACC-CCCUACUCGUAGU-----ACGAAGAUGCCUUCUUAAUAUGUGGUCGCUGC--UCGCCCACACAGUGAGU-------- .....................-....(((((..((-----..((((....)))).......((((.((....--.)).))))))..)))))-------- ( -14.30) >consensus CUGGAUUCAG_____UGCACCACCCUGGUGGUGCUGCUUCACUGUGGUGCAUGCUUAAUAUGUGGUCGCUGCCAUCGCCCACACAGUGAGUCGCAUCCC ................(((((((....)))))))...........(((((..(((((...(((((.((.......)).)))))...))))).))))).. (-22.14 = -23.87 + 1.72)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:28:16 2006