| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 15,555,661 – 15,555,786 |
| Length | 125 |
| Max. P | 0.801101 |

| Location | 15,555,661 – 15,555,754 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 80.48 |
| Mean single sequence MFE | -23.43 |
| Consensus MFE | -16.04 |
| Energy contribution | -16.43 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.547051 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15555661 93 + 20766785 G-------AGAACAU-UCGGCGCAGAAUGGAGAACGAGAACGAGGC---ACCUCAGUAACUUGUCAGAAUUGUGACAAGUUGUUAUCGAAAUUCAGGGGAUCUC (-------(((.(((-((......)))))(((..(((.((((((..---..)))...(((((((((......)))))))))))).)))...)))......)))) ( -26.90) >DroPse_CAF1 388723 102 + 1 GAGAACAGAGAACAC-UGAACACAGAGCGGCAAUGGAGAACGCUGCACCACUUCAGUAACUUGUCAGAAUUGUGACAAGUUGUUAUCGAAAUUUCAGAGUUCU- ........(((((.(-((((....(((((((..........)))))..........((((((((((......))))))))))...)).....))))).)))))- ( -28.70) >DroSec_CAF1 349252 93 + 1 G-------AGAACAU-UCGGCGCAGAAUGGAGAACGAGAACGAGGC---ACUUCAGUAACUUGUCAAAAUUGUGACAAGUUGUUAUCGAAAUUCAGGGGAUCUC (-------(((.(((-((......)))))(((..(((.((((((..---..)))...(((((((((......)))))))))))).)))...)))......)))) ( -23.70) >DroSim_CAF1 329157 93 + 1 G-------AGAACAU-UCGGCGCAGAAUGGAGAACGAGAACGAGGC---ACUUCAGUAACUUGUCAGAAUUGUGACAAGUUGUUAUCGAAAUUCGGGGGAUCUC (-------(((.(((-((......)))))(((..(((.((((((..---..)))...(((((((((......)))))))))))).)))...)))......)))) ( -23.60) >DroEre_CAF1 337114 72 + 1 C-------AGA----------------------GCGAGAACGAGGC---ACUUCAGUAACUUGUCAGAAUUGUGACAAGUUGUUAUCGAAAUUCAGGGGAUCUG (-------(((----------------------.(((.((((((..---..)))...(((((((((......)))))))))))).)))....((....)))))) ( -16.30) >DroYak_CAF1 348649 94 + 1 G-------AGAACAUUUCGGCGGAGAAUGGUGAACGAGAACGAGGC---ACUUCAGUAACUUGUCAAAAUUGUGACAAGUUGUUAUCGAAAUUCAGGGGAUCUC (-------(((..(((((((..(.((..((((..((....))...)---)))))..((((((((((......)))))))))))..)))))))((....)))))) ( -21.40) >consensus G_______AGAACAU_UCGGCGCAGAAUGGAGAACGAGAACGAGGC___ACUUCAGUAACUUGUCAGAAUUGUGACAAGUUGUUAUCGAAAUUCAGGGGAUCUC ..................................(((.(((((((.....))))...(((((((((......)))))))))))).)))................ (-16.04 = -16.43 + 0.39)

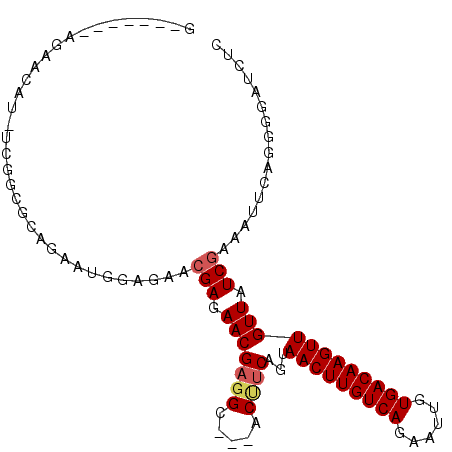
| Location | 15,555,661 – 15,555,754 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 80.48 |
| Mean single sequence MFE | -21.29 |
| Consensus MFE | -13.93 |
| Energy contribution | -14.12 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.801101 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15555661 93 - 20766785 GAGAUCCCCUGAAUUUCGAUAACAACUUGUCACAAUUCUGACAAGUUACUGAGGU---GCCUCGUUCUCGUUCUCCAUUCUGCGCCGA-AUGUUCU-------C ((((......(((...(((.((((((((((((......)))))))))...(((..---..)))))).))))))..(((((......))-))).)))-------) ( -24.10) >DroPse_CAF1 388723 102 - 1 -AGAACUCUGAAAUUUCGAUAACAACUUGUCACAAUUCUGACAAGUUACUGAAGUGGUGCAGCGUUCUCCAUUGCCGCUCUGUGUUCA-GUGUUCUCUGUUCUC -(((((...(((...........(((((((((......)))))))))(((((((..(.((.(((........))).)).)..).))))-)).)))...))))). ( -29.60) >DroSec_CAF1 349252 93 - 1 GAGAUCCCCUGAAUUUCGAUAACAACUUGUCACAAUUUUGACAAGUUACUGAAGU---GCCUCGUUCUCGUUCUCCAUUCUGCGCCGA-AUGUUCU-------C ((((........((((((.....(((((((((......)))))))))..))))))---.......))))......(((((......))-)))....-------. ( -20.26) >DroSim_CAF1 329157 93 - 1 GAGAUCCCCCGAAUUUCGAUAACAACUUGUCACAAUUCUGACAAGUUACUGAAGU---GCCUCGUUCUCGUUCUCCAUUCUGCGCCGA-AUGUUCU-------C ((((........((((((.....(((((((((......)))))))))..))))))---.......))))......(((((......))-)))....-------. ( -20.26) >DroEre_CAF1 337114 72 - 1 CAGAUCCCCUGAAUUUCGAUAACAACUUGUCACAAUUCUGACAAGUUACUGAAGU---GCCUCGUUCUCGC----------------------UCU-------G (((.....))).....(((.((((((((((((......)))))))))...((...---...))))).))).----------------------...-------. ( -14.70) >DroYak_CAF1 348649 94 - 1 GAGAUCCCCUGAAUUUCGAUAACAACUUGUCACAAUUUUGACAAGUUACUGAAGU---GCCUCGUUCUCGUUCACCAUUCUCCGCCGAAAUGUUCU-------C ((((.....((((...(((.((((((((((((......)))))))))...((...---...))))).)))))))....))))..............-------. ( -18.80) >consensus GAGAUCCCCUGAAUUUCGAUAACAACUUGUCACAAUUCUGACAAGUUACUGAAGU___GCCUCGUUCUCGUUCUCCAUUCUGCGCCGA_AUGUUCU_______C ((((........((((((.....(((((((((......)))))))))..))))))..........))))................................... (-13.93 = -14.12 + 0.19)


| Location | 15,555,679 – 15,555,786 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 83.37 |
| Mean single sequence MFE | -24.81 |
| Consensus MFE | -15.36 |
| Energy contribution | -16.39 |
| Covariance contribution | 1.03 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.62 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.520767 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15555679 107 - 20766785 CCCUCCCUGGCUCGCCAGCUCUUCAG--------AUCCCAGAGAUCCCCUGAAUUUCGAUAACAACUUGUCACAAUUCUGACAAGUUACUGAGGU---GCCUCGUUCUCGUUCUCCAU ......((((....))))...(((((--------(((.....))))...))))...(((.((((((((((((......)))))))))...(((..---..)))))).)))........ ( -26.20) >DroPse_CAF1 388748 101 - 1 CCCUCUCGA----GCCAC--AUCCAG--------AUCC---AGAACUCUGAAAUUUCGAUAACAACUUGUCACAAUUCUGACAAGUUACUGAAGUGGUGCAGCGUUCUCCAUUGCCGC .........----(((((--...(((--------...(---((....))).............(((((((((......))))))))).)))..)))))((.(((........))).)) ( -25.10) >DroSec_CAF1 349270 107 - 1 CCCUCCCUGGCUCGACAGCUCUCCAG--------AUCCCAGAGAUCCCCUGAAUUUCGAUAACAACUUGUCACAAUUUUGACAAGUUACUGAAGU---GCCUCGUUCUCGUUCUCCAU ........(((.(..(((.((....(--------(((.....))))....))...........(((((((((......))))))))).)))..).---)))................. ( -21.60) >DroSim_CAF1 329175 107 - 1 CCCUCCCUGGCUCGCCAGCUCUCCAG--------AUCCGAGAGAUCCCCCGAAUUUCGAUAACAACUUGUCACAAUUCUGACAAGUUACUGAAGU---GCCUCGUUCUCGUUCUCCAU ......((((....))))......((--------(..((((((.........((((((.....(((((((((......)))))))))..))))))---......)))))).))).... ( -24.46) >DroEre_CAF1 337118 100 - 1 CCCUCUCUGGCUCGCCAGCUCUCCAG--------AUCCCGCAGAUCCCCUGAAUUUCGAUAACAACUUGUCACAAUUCUGACAAGUUACUGAAGU---GCCUCGUUCUCGC------- .......(((....)))((.((.(((--------......(((.....)))............(((((((((......))))))))).))).)).---))...........------- ( -21.10) >DroYak_CAF1 348668 115 - 1 CCUUCUCUGGCUCGCCAGCUCUCCAGUUCUCUGGAUCCCAGAGAUCCCCUGAAUUUCGAUAACAACUUGUCACAAUUUUGACAAGUUACUGAAGU---GCCUCGUUCUCGUUCACCAU ...(((((((((....)))..(((((....)))))...)))))).....((((...(((.((((((((((((......)))))))))...((...---...))))).))))))).... ( -30.40) >consensus CCCUCCCUGGCUCGCCAGCUCUCCAG________AUCCCAGAGAUCCCCUGAAUUUCGAUAACAACUUGUCACAAUUCUGACAAGUUACUGAAGU___GCCUCGUUCUCGUUCUCCAU ......((((....))))......................((((........((((((.....(((((((((......)))))))))..))))))..........))))......... (-15.36 = -16.39 + 1.03)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:28:12 2006