| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 15,549,801 – 15,549,895 |
| Length | 94 |
| Max. P | 0.825576 |

| Location | 15,549,801 – 15,549,895 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 78.92 |
| Mean single sequence MFE | -29.09 |
| Consensus MFE | -18.05 |
| Energy contribution | -18.77 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.797404 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
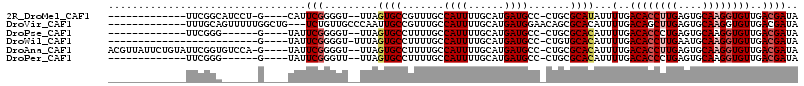
>2R_DroMel_CAF1 15549801 94 + 20766785 -------------UUCGGCAUCCU-G----CAUUCGGGGU--UUAGUGCCGUUUGCCAUUUUGCAUGAUGCC-CUGCGCAUAUUUUGACACCUUGAGUGCAAGGUGUUGACGAUA -------------.((((((((.(-(----((((((((((--(((((((.((..(.((((......)))).)-..))))))....))).)))))))))))).))))))))..... ( -33.50) >DroVir_CAF1 469365 99 + 1 -------------UUUGCAGUUUUUGGCUG---UCUGUUGCCCAAUUGCCGUUUGCCAUUUUGCAUGAUGGAACAGCGCACAUUUUGACAGCUUGAGUGCAAGGUGUUGACGAUA -------------((((((.((...(((((---((.(((((.......((((((((......))).)))))......))).))...))))))).)).))))))............ ( -27.62) >DroPse_CAF1 382448 89 + 1 -------------UUCGGG------G----UAUUCGGGGU--UUAGUGCCUUUUGCCAUUUUGCAUGAUGCC-CUGCGCACAUUUUGACACCCUGAGUGCAAGGUGUUGACGAUA -------------..((((------(----((((.(((((--.....))))).(((......))).))))))-)))(((((((((((.(((.....)))))))))).)).))... ( -27.10) >DroWil_CAF1 636856 84 + 1 -------------------------G----UAUUCGGGGU-UUUAGUGCCUUUUGCCAUUUUGCAUGAUGCC-CUGUGCACAUUUUGACACCUUGAAUGCAAGGUGUUGACGAUA -------------------------(----((..((((((-....((((.............))))...)))-)))))).....(..((((((((....))))))))..)..... ( -26.82) >DroAna_CAF1 318632 107 + 1 ACGUUAUUCUGUAUUCGGUGUCCA-G----UAUUCGGGGU--UUAGUGCCUUUUGCCAUUUUGCAUGAUGCC-CUGCGCACAUUUUGACACCUUGAGUGCAAGGUGUUGACGAUA .(((((.(((((((((((((((..-(----(...((((((--...((((.............))))...)))-)))...)).....))))))..)))))).)))...)))))... ( -32.72) >DroPer_CAF1 347563 89 + 1 -------------UUCGGG------G----UAUUCGGGUU--UUAGUGCCUUUUGCCAUUUUGCAUGAUGCC-CUGCGCACAUUUUGACACCCUGAGUGCAAGGUGUUGACGAUA -------------..((((------(----((((.((((.--.....))))..(((......))).))))))-)))(((((((((((.(((.....)))))))))).)).))... ( -26.80) >consensus _____________UUCGGG______G____UAUUCGGGGU__UUAGUGCCUUUUGCCAUUUUGCAUGAUGCC_CUGCGCACAUUUUGACACCUUGAGUGCAAGGUGUUGACGAUA .................................(((.........((((.......((((......)))).......))))...(..((((((((....))))))))..)))).. (-18.05 = -18.77 + 0.72)
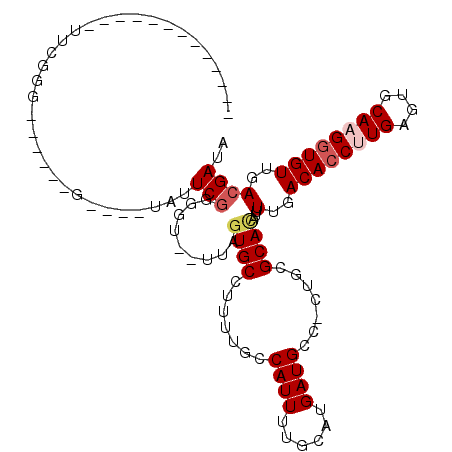
| Location | 15,549,801 – 15,549,895 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 78.92 |
| Mean single sequence MFE | -21.58 |
| Consensus MFE | -16.66 |
| Energy contribution | -16.93 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.825576 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15549801 94 - 20766785 UAUCGUCAACACCUUGCACUCAAGGUGUCAAAAUAUGCGCAG-GGCAUCAUGCAAAAUGGCAAACGGCACUAA--ACCCCGAAUG----C-AGGAUGCCGAA------------- ..((....((((((((....))))))))..............-((((((.((((...(((.....((......--.)))))..))----)-).)))))))).------------- ( -26.10) >DroVir_CAF1 469365 99 - 1 UAUCGUCAACACCUUGCACUCAAGCUGUCAAAAUGUGCGCUGUUCCAUCAUGCAAAAUGGCAAACGGCAAUUGGGCAACAGA---CAGCCAAAAACUGCAAA------------- .............(((((.....((((((.........((((((((((........))))..)))))).....(....).))---)))).......))))).------------- ( -24.80) >DroPse_CAF1 382448 89 - 1 UAUCGUCAACACCUUGCACUCAGGGUGUCAAAAUGUGCGCAG-GGCAUCAUGCAAAAUGGCAAAAGGCACUAA--ACCCCGAAUA----C------CCCGAA------------- ..(((...((((((((....))))))))......((((((..-.))....(((......)))....))))...--....)))...----.------......------------- ( -18.90) >DroWil_CAF1 636856 84 - 1 UAUCGUCAACACCUUGCAUUCAAGGUGUCAAAAUGUGCACAG-GGCAUCAUGCAAAAUGGCAAAAGGCACUAAA-ACCCCGAAUA----C------------------------- ..(((...((((((((....))))))))......((((....-.((..(((.....))))).....))))....-....)))...----.------------------------- ( -19.60) >DroAna_CAF1 318632 107 - 1 UAUCGUCAACACCUUGCACUCAAGGUGUCAAAAUGUGCGCAG-GGCAUCAUGCAAAAUGGCAAAAGGCACUAA--ACCCCGAAUA----C-UGGACACCGAAUACAGAAUAACGU ....(((.((((((((....))))))))......((((((..-.))....(((......)))....))))...--..........----.-..)))................... ( -21.20) >DroPer_CAF1 347563 89 - 1 UAUCGUCAACACCUUGCACUCAGGGUGUCAAAAUGUGCGCAG-GGCAUCAUGCAAAAUGGCAAAAGGCACUAA--AACCCGAAUA----C------CCCGAA------------- ..(((...((((((((....))))))))......((((((..-.))....(((......)))....))))...--....)))...----.------......------------- ( -18.90) >consensus UAUCGUCAACACCUUGCACUCAAGGUGUCAAAAUGUGCGCAG_GGCAUCAUGCAAAAUGGCAAAAGGCACUAA__ACCCCGAAUA____C______CCCGAA_____________ ....(((.((((((((....)))))))).......(((.((...((.....))....)))))...)))............................................... (-16.66 = -16.93 + 0.28)

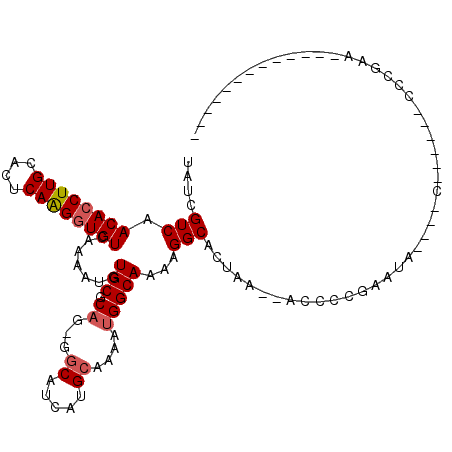
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:28:08 2006