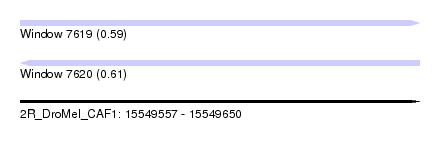
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 15,549,557 – 15,549,650 |
| Length | 93 |
| Max. P | 0.608863 |

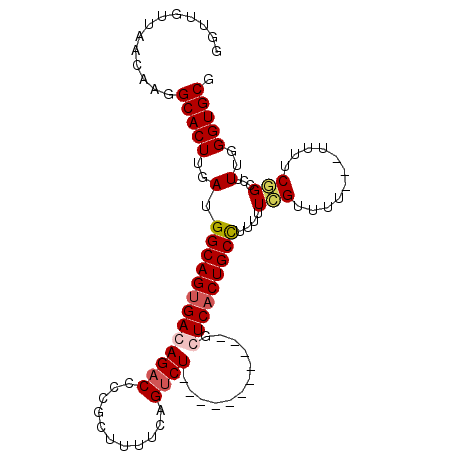
| Location | 15,549,557 – 15,549,650 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 83.84 |
| Mean single sequence MFE | -26.38 |
| Consensus MFE | -14.75 |
| Energy contribution | -16.00 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.592352 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
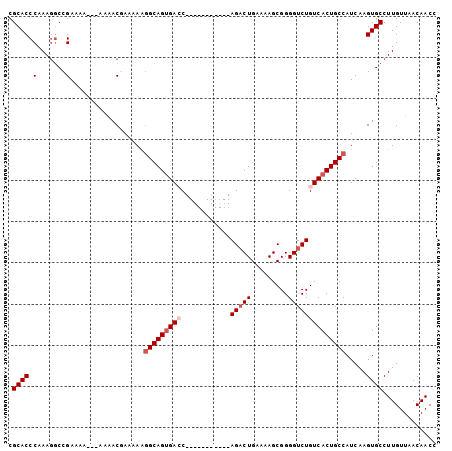
>2R_DroMel_CAF1 15549557 93 + 20766785 CGCACCCAAAGGCCGAAAAAAAAAAACGAAAAAGGCAGUGACC-----------AGACUGAAAAGCGGGGUCUGUCACUGCCAUCAAGUGCCUUGUUAACAACC ........(((((((...........)).....(((((((((.-----------(((((.........)))))))))))))).......))))).......... ( -27.40) >DroSec_CAF1 343288 90 + 1 CGCACCCAAAGGCCGAAAA---AAAACGAAAAAGGCAGUGAGC-----------AGACUGAAAAGCGGGGUCUGUCACUGCCAUCAAGUGCCUUGUUAACAACC ........(((((((....---....)).....((((((((.(-----------(((((.........)))))))))))))).......))))).......... ( -28.40) >DroSim_CAF1 323148 90 + 1 CGCACCCAAAGGCCGAAAA---AAAACUAAAAAGGCAGUGAGC-----------AGACUGAAAAGCGGGGUCUGUCACUGCCAUCAAGUGCCUUGUUAACAACC ........(((((((....---....)......((((((((.(-----------(((((.........)))))))))))))).....).))))).......... ( -26.70) >DroAna_CAF1 318383 104 + 1 CGCACCCAAAAACAGAGUCCCAGAGACCAUAAGAGCAGAGACCCUCGGAGCCAGAGUCUGAAAAGCGGGGUCUGUCACUGCCAUCAAGUGCCUUGUUAACAACC .((((.........(.(((.....))))....(.((((.(((....(((.((...(.((....))).)).)))))).))))).....))))............. ( -23.00) >consensus CGCACCCAAAGGCCGAAAA___AAAACGAAAAAGGCAGUGACC___________AGACUGAAAAGCGGGGUCUGUCACUGCCAUCAAGUGCCUUGUUAACAACC .((((.(.......)..................(((((((((............(((((.........)))))))))))))).....))))............. (-14.75 = -16.00 + 1.25)

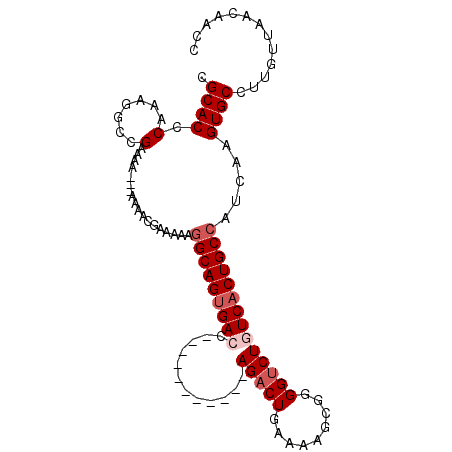
| Location | 15,549,557 – 15,549,650 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 83.84 |
| Mean single sequence MFE | -30.16 |
| Consensus MFE | -19.11 |
| Energy contribution | -19.93 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.608863 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15549557 93 - 20766785 GGUUGUUAACAAGGCACUUGAUGGCAGUGACAGACCCCGCUUUUCAGUCU-----------GGUCACUGCCUUUUUCGUUUUUUUUUUUCGGCCUUUGGGUGCG .............(((((..(.(((((((((((((...........))))-----------.)))))))))....(((...........)))...)..))))). ( -29.50) >DroSec_CAF1 343288 90 - 1 GGUUGUUAACAAGGCACUUGAUGGCAGUGACAGACCCCGCUUUUCAGUCU-----------GCUCACUGCCUUUUUCGUUUU---UUUUCGGCCUUUGGGUGCG .............(((((..(.(((((((((((((...........))))-----------).))))))))....(((....---....)))...)..))))). ( -30.50) >DroSim_CAF1 323148 90 - 1 GGUUGUUAACAAGGCACUUGAUGGCAGUGACAGACCCCGCUUUUCAGUCU-----------GCUCACUGCCUUUUUAGUUUU---UUUUCGGCCUUUGGGUGCG .............(((((..(.(((((((((((((...........))))-----------).))))))))......(((..---.....)))..)..))))). ( -28.70) >DroAna_CAF1 318383 104 - 1 GGUUGUUAACAAGGCACUUGAUGGCAGUGACAGACCCCGCUUUUCAGACUCUGGCUCCGAGGGUCUCUGCUCUUAUGGUCUCUGGGACUCUGUUUUUGGGUGCG .............(((((..(..((((.(.((((..(((......((((((((....).))))))).........)))..))))...).))))..)..))))). ( -31.96) >consensus GGUUGUUAACAAGGCACUUGAUGGCAGUGACAGACCCCGCUUUUCAGUCU___________GCUCACUGCCUUUUUCGUUUU___UUUUCGGCCUUUGGGUGCG .............(((((..(.(((((((((((((...........))))............)))))))))....(((...........)))...)..))))). (-19.11 = -19.93 + 0.81)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:28:06 2006