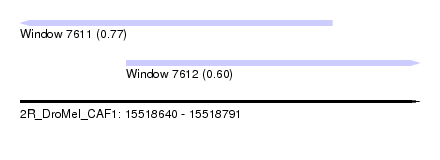
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 15,518,640 – 15,518,791 |
| Length | 151 |
| Max. P | 0.766114 |

| Location | 15,518,640 – 15,518,758 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.16 |
| Mean single sequence MFE | -30.90 |
| Consensus MFE | -28.67 |
| Energy contribution | -28.42 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.766114 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15518640 118 - 20766785 UCA--AAUAGGUCUGUUAGCCCAUCUCUUCAAGCCCGUUAUAAUAUACGACGGCACUGAAGUGAGCUCAGACUUCUCAAGUGUCUAUAAAGAUACCGAAGAGGGCAUUCGAUUCCAGAAA ...--....((..((...((((.((.(((((.((((((........)))..)))..))))).)).......((((....((((((....)))))).)))).))))...))...))..... ( -29.50) >DroSec_CAF1 312167 118 - 1 UCA--AAAAGGUCUGUUGGCCCAUCUCUUCAAGCCCGUUAUAAUAUACGACGGUACUGAAGUGAGCUCAGACUUCUCAAGUGUCUACAAAGAUACCGAAGAGGGCAUUCGAUUCCAGAAA ...--....((..((...((((.((.(((((...((((..((...))..))))...))))).)).......((((....((((((....)))))).)))).))))...))...))..... ( -30.50) >DroSim_CAF1 292075 118 - 1 UCA--AAAAGGUCUGUUGGCCCAUCUCUUCAAGCCCGUUAUAAUAUACGACGGUACCGAAGUGAGCUCAGACUUCUCAGGUGUCUACAAAGAUACCGAAGAGGGCAUUCGAUUCCAGAAA ...--....((..((...((((...(((((....((((..((...))..))))....(((((........)))))...(((((((....))))))))))))))))...))...))..... ( -32.80) >DroYak_CAF1 309875 120 - 1 UAAACAAUAGGUUUAUUGGCCCAUCUCUUCAAACCUGUUAUUAUAUACGACGGUACUGAAGUGACCUCAGACUUCUCAAGUGUCUAUAAAGAUACCGAGGAGGGCAUUCGGUUCCAGAAU .........((..(....((((.((.(((((...((((..(.....)..))))...))))).))((((.((....))..((((((....)))))).)))).)))).....)..))..... ( -30.80) >consensus UCA__AAAAGGUCUGUUGGCCCAUCUCUUCAAGCCCGUUAUAAUAUACGACGGUACUGAAGUGAGCUCAGACUUCUCAAGUGUCUACAAAGAUACCGAAGAGGGCAUUCGAUUCCAGAAA .........((...((((((((.((.(((((...((((..(.....)..))))...))))).)).......((((....((((((....)))))).)))).))))...)))).))..... (-28.67 = -28.42 + -0.25)


| Location | 15,518,680 – 15,518,791 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 86.05 |
| Mean single sequence MFE | -29.12 |
| Consensus MFE | -19.39 |
| Energy contribution | -19.57 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.601239 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15518680 111 + 20766785 CUUGAGAAGUCUGAGCUCACUUCAGUGCCGUCGUAUAUUAUAACGGGCUUGAAGAGAUGGGCUAACAGACCUAUU--UGAAAAAGGAAAAGGGUU--U--AGUAUAAGCUUUAUGGA .....(((((((((((((.((((((.(((..(((........))))))))))))((((((((.....).))))))--)............)))))--)--)).....)))))..... ( -28.30) >DroSec_CAF1 312207 110 + 1 CUUGAGAAGUCUGAGCUCACUUCAGUACCGUCGUAUAUUAUAACGGGCUUGAAGAGAUGGGCCAACAGACCUUUU--UGAA-AAGAAAAAGGGUU--U--AGUAUAGGCUUUAUGGA .....((((((((.(((((((((((..((((..((...))..))))..))))))...)))))..(((((((((((--(...-....)))))))))--)--.)).))))))))..... ( -34.00) >DroSim_CAF1 292115 110 + 1 CCUGAGAAGUCUGAGCUCACUUCGGUACCGUCGUAUAUUAUAACGGGCUUGAAGAGAUGGGCCAACAGACCUUUU--UGAA-AAGGAAAAGGGUU--U--AGUAUAAGCUUUAUGGC ((...(((((.((.(((((((((((..((((..((...))..))))..))))))...)))))))(((((((((((--(...-....)))))))))--)--.))....)))))..)). ( -29.10) >DroYak_CAF1 309915 113 + 1 CUUGAGAAGUCUGAGGUCACUUCAGUACCGUCGUAUAUAAUAACAGGUUUGAAGAGAUGGGCCAAUAAACCUAUUGUUUAA----AAAAUGUGUUAUUAGAAUAUAAGCUUUACAUA ..((.(((((((((((...)))))).......((((.((((((((.((((.(((.((((((........)))))).)))..----.)))).))))))))..))))..))))).)).. ( -25.10) >consensus CUUGAGAAGUCUGAGCUCACUUCAGUACCGUCGUAUAUUAUAACGGGCUUGAAGAGAUGGGCCAACAGACCUAUU__UGAA_AAGAAAAAGGGUU__U__AGUAUAAGCUUUAUGGA .....(((((.((.(((((((((((..((((..(.....)..))))..))))))...)))))))...((((((((............))))))))............)))))..... (-19.39 = -19.57 + 0.19)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:27:58 2006