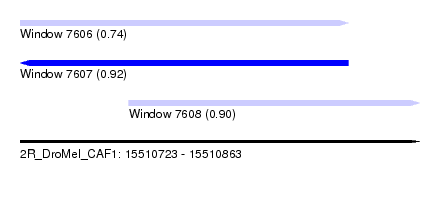
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 15,510,723 – 15,510,863 |
| Length | 140 |
| Max. P | 0.916866 |

| Location | 15,510,723 – 15,510,838 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.96 |
| Mean single sequence MFE | -27.63 |
| Consensus MFE | -24.03 |
| Energy contribution | -24.20 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.743645 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
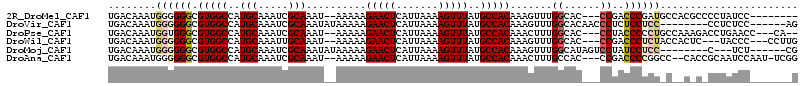
>2R_DroMel_CAF1 15510723 115 + 20766785 AAC-U-UAAUGCAAUUUCGGUCUGGCAGAUUUCAUUUGUGUGACAAAUGGGGGGCGUGGCCAUGCAAAUCGCAAAU--AAAAAGAACUCAUUAAAAGUUUAUGCCACAAAGUUUGGCAC- ...-(-(((((....(((((((..((...(..((((((.....))))))..).))..)))).(((.....)))...--.....)))..)))))).......(((((.......))))).- ( -28.30) >DroVir_CAF1 414654 119 + 1 AACUU-UAAUGCAAUUUCGCUCUAGCAGAUUUCAUUUGUGUGACAAAUGGGGGGCGUGGCCAUGCAAAUCGCAAAUAUAAAAAGAACUCAUUAAAAGUUUAUGCCACAAAGUUUGGCACA (((((-(((((....((((((...((...(..((((((.....))))))..).))..)))..(((.....)))..........)))..)))).))))))..(((((.......))))).. ( -26.40) >DroPse_CAF1 343307 115 + 1 AAC-U-UAAUGCAAUUUCGCUCCAGCAGAUUUCAUUUGUGUGACAAAUGGUGGGCGUGGCCAUGCAAAUCGCAAAU--AAAAAGAACUCAUUAAAAGUUUAUGCCACAAACUUUGGCAC- ...-.-...(((......((.(((.((....((((....))))....)).)))))(((((..(((.....)))...--.....(((((.......)))))..)))))........))).- ( -27.30) >DroWil_CAF1 574318 116 + 1 AAC-UUUAAUGCAAUUUCGCUCAACCAGAUUUCAUUUGUGUGACAAAUGGGGGGCGUGGCCAUGCAAAUUGCAAAU--AAAAAGAACUCAUUAAAAGUUUAUGCCACAAAGUUUGGCAC- ...-.....((((((((.((....((((.(..((((((.....))))))..)..).)))....))))))))))...--.....(((((.......))))).(((((.......))))).- ( -28.30) >DroMoj_CAF1 467601 119 + 1 AACUU-UAAUGCAAUUUCGCUCUAGCAGAUUUCAUUUGUGUGACAAAUGGGGGGCGUGGCCAUGCAAAUCGCAAAUAUAAAAAGAACUCAUUAAAAGUUUAUGCCACAAAGUUUGGCAUA (((((-(((((....((((((...((...(..((((((.....))))))..).))..)))..(((.....)))..........)))..)))).))))))(((((((.......))))))) ( -28.20) >DroPer_CAF1 306937 115 + 1 AAC-U-UAAUGCAAUUUCGCUCCAGCAGAUUUCAUUUGUGUGACAAAUGGUGGGCGUGGCCAUGCAAAUCGCAAAU--AAAAAGAACUCAUUAAAAGUUUAUGCCACAAACUUUGGCAC- ...-.-...(((......((.(((.((....((((....))))....)).)))))(((((..(((.....)))...--.....(((((.......)))))..)))))........))).- ( -27.30) >consensus AAC_U_UAAUGCAAUUUCGCUCUAGCAGAUUUCAUUUGUGUGACAAAUGGGGGGCGUGGCCAUGCAAAUCGCAAAU__AAAAAGAACUCAUUAAAAGUUUAUGCCACAAAGUUUGGCAC_ .........(((............((...(..((((((.....))))))..).))(((((..(((.....)))..........(((((.......)))))..)))))........))).. (-24.03 = -24.20 + 0.17)

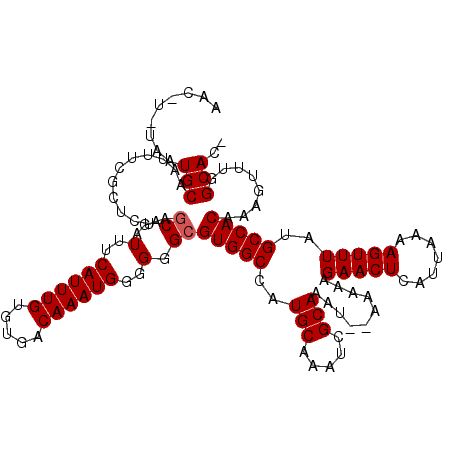
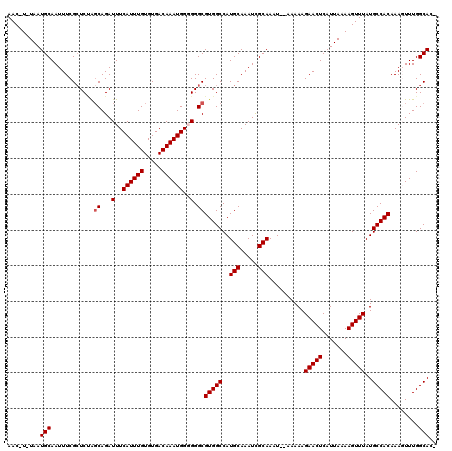
| Location | 15,510,723 – 15,510,838 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.96 |
| Mean single sequence MFE | -26.68 |
| Consensus MFE | -24.71 |
| Energy contribution | -24.60 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.916866 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15510723 115 - 20766785 -GUGCCAAACUUUGUGGCAUAAACUUUUAAUGAGUUCUUUUU--AUUUGCGAUUUGCAUGGCCACGCCCCCCAUUUGUCACACAAAUGAAAUCUGCCAGACCGAAAUUGCAUUA-A-GUU -((((((.......)))))).(((((.....)))))...(((--(..(((((((((..((((.........((((((.....))))))......))))...).)))))))).))-)-).. ( -24.16) >DroVir_CAF1 414654 119 - 1 UGUGCCAAACUUUGUGGCAUAAACUUUUAAUGAGUUCUUUUUAUAUUUGCGAUUUGCAUGGCCACGCCCCCCAUUUGUCACACAAAUGAAAUCUGCUAGAGCGAAAUUGCAUUA-AAGUU (((((((.......)))))))(((((.....)))))..(((((....((((((((((..(((...)))...((((((.....))))))............)).)))))))).))-))).. ( -27.00) >DroPse_CAF1 343307 115 - 1 -GUGCCAAAGUUUGUGGCAUAAACUUUUAAUGAGUUCUUUUU--AUUUGCGAUUUGCAUGGCCACGCCCACCAUUUGUCACACAAAUGAAAUCUGCUGGAGCGAAAUUGCAUUA-A-GUU -((((((.......)))))).(((((.....)))))...(((--(..((((((((((....(((.((....((((((.....))))))......))))).)).)))))))).))-)-).. ( -26.50) >DroWil_CAF1 574318 116 - 1 -GUGCCAAACUUUGUGGCAUAAACUUUUAAUGAGUUCUUUUU--AUUUGCAAUUUGCAUGGCCACGCCCCCCAUUUGUCACACAAAUGAAAUCUGGUUGAGCGAAAUUGCAUUAAA-GUU -((((((.......)))))).(((((.....)))))..((((--(..((((((((((.((((((.......((((((.....)))))).....)))))).)).)))))))).))))-).. ( -28.90) >DroMoj_CAF1 467601 119 - 1 UAUGCCAAACUUUGUGGCAUAAACUUUUAAUGAGUUCUUUUUAUAUUUGCGAUUUGCAUGGCCACGCCCCCCAUUUGUCACACAAAUGAAAUCUGCUAGAGCGAAAUUGCAUUA-AAGUU (((((((.......)))))))(((((.....)))))..(((((....((((((((((..(((...)))...((((((.....))))))............)).)))))))).))-))).. ( -27.00) >DroPer_CAF1 306937 115 - 1 -GUGCCAAAGUUUGUGGCAUAAACUUUUAAUGAGUUCUUUUU--AUUUGCGAUUUGCAUGGCCACGCCCACCAUUUGUCACACAAAUGAAAUCUGCUGGAGCGAAAUUGCAUUA-A-GUU -((((((.......)))))).(((((.....)))))...(((--(..((((((((((....(((.((....((((((.....))))))......))))).)).)))))))).))-)-).. ( -26.50) >consensus _GUGCCAAACUUUGUGGCAUAAACUUUUAAUGAGUUCUUUUU__AUUUGCGAUUUGCAUGGCCACGCCCCCCAUUUGUCACACAAAUGAAAUCUGCUAGAGCGAAAUUGCAUUA_A_GUU .((((((.......)))))).(((((.....)))))...........((((((((((..(((...)))...((((((.....))))))............)).))))))))......... (-24.71 = -24.60 + -0.11)

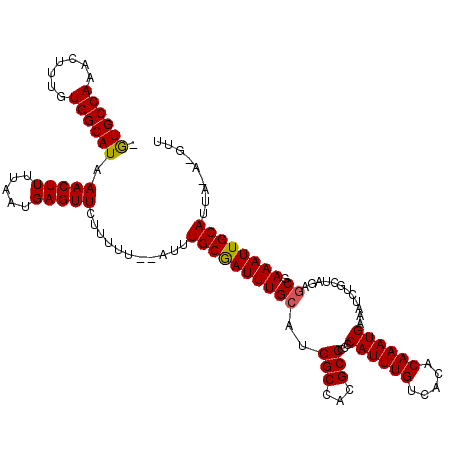
| Location | 15,510,761 – 15,510,863 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 79.58 |
| Mean single sequence MFE | -26.51 |
| Consensus MFE | -17.70 |
| Energy contribution | -18.23 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.898897 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
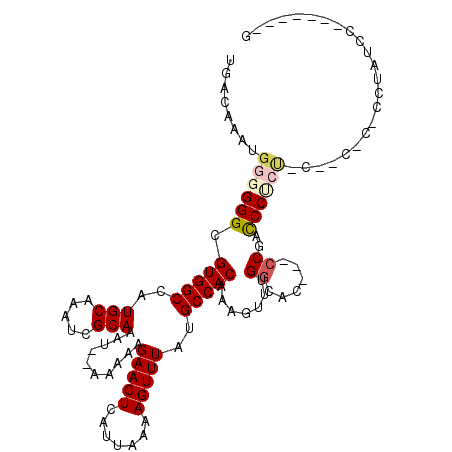
>2R_DroMel_CAF1 15510761 102 + 20766785 UGACAAAUGGGGGGCGUGGCCAUGCAAAUCGCAAAU--AAAAAGAACUCAUUAAAAGUUUAUGCCACAAAGUUUGGCAC---CCGACCCGAUGCCACGCCCCUAUCC-------- .........(((((((((((..(((.....)))...--.....(((((.......))))).(((((.......))))).---..........)))))))))))....-------- ( -34.00) >DroVir_CAF1 414693 101 + 1 UGACAAAUGGGGGGCGUGGCCAUGCAAAUCGCAAAUAUAAAAAGAACUCAUUAAAAGUUUAUGCCACAAAGUUUGGCACAACCCUCUCCUCC--------CCUCUCC------AG ........((((((.(((((..(((.....)))..........(((((.......)))))..))))).......((......))....))))--------)).....------.. ( -23.10) >DroPse_CAF1 343345 105 + 1 UGACAAAUGGUGGGCGUGGCCAUGCAAAUCGCAAAU--AAAAAGAACUCAUUAAAAGUUUAUGCCACAAACUUUGGCAC---CCUACCCCCUGCCAAAGACCUGAACC---CA-- ..........((((.(((((..(((.....)))...--.....(((((.......)))))..)))))...((((((((.---.........)))))))).......))---))-- ( -27.30) >DroWil_CAF1 574357 104 + 1 UGACAAAUGGGGGGCGUGGCCAUGCAAAUUGCAAAU--AAAAAGAACUCAUUAAAAGUUUAUGCCACAAAGUUUGGCAC---CCGACCCUCUACCACUC---UACCC---CCUUG ........(((((..(((((..(((.....)))...--.....(((((.......)))))..)))))..((..(((...---...........)))..)---).)))---))... ( -25.84) >DroMoj_CAF1 467640 98 + 1 UGACAAAUGGGGGGCGUGGCCAUGCAAAUCGCAAAUAUAAAAAGAACUCAUUAAAAGUUUAUGCCACAAAGUUUGGCAUAGUCCUAUCCUCC--------C---UCU------CG ........((((((...((.(.(((.....)))..........................(((((((.......)))))))).))...)))))--------)---...------.. ( -23.70) >DroAna_CAF1 282457 107 + 1 UGACAAAUGGGGGGCGUGGCCAUGCAAAUCGCAAAU--AAAAAGAACUCAUUAAAAGUUUAUGCCACAAACUUUGCCAC---CCGACCCCGGCC--CACCGCAAUCCAAU-UCGG .......(((((((((((((..(((.....)))...--...............(((((((.......))))))))))))---.((....)))))--).)).)).......-.... ( -25.10) >consensus UGACAAAUGGGGGGCGUGGCCAUGCAAAUCGCAAAU__AAAAAGAACUCAUUAAAAGUUUAUGCCACAAAGUUUGGCAC___CCGACCCUCU_C__C_C_CCUAUCC_______G ........((((((.(((((..(((.....)))..........(((((.......)))))..))))).......((......))..))))))....................... (-17.70 = -18.23 + 0.53)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:27:54 2006