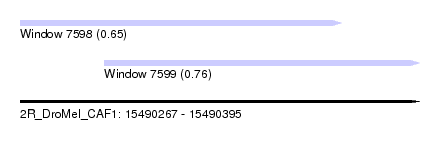
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 15,490,267 – 15,490,395 |
| Length | 128 |
| Max. P | 0.755630 |

| Location | 15,490,267 – 15,490,370 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 76.86 |
| Mean single sequence MFE | -20.11 |
| Consensus MFE | -12.60 |
| Energy contribution | -12.60 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.651100 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
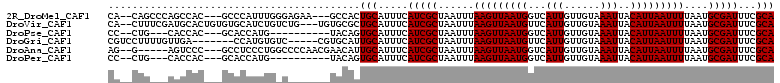
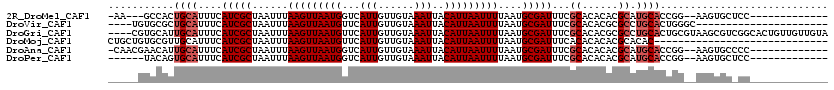
>2R_DroMel_CAF1 15490267 103 + 20766785 CA--CAGCCCAGCCAC---GCCCAUUUGGGAGAA---GCCACUGCAUUUCAUCGCUAAUUUAAGUUAAUGGUCAUUGUUGUAAAUUACAUUAAUUUUAAUGCGAUUUCGCA ..--..((...((..(---.(((....))).)..---))...........(((((......(((((((((...(((......)))..)))))))))....)))))...)). ( -19.50) >DroVir_CAF1 381392 106 + 1 CA--CUUUCGAUGCACUGUGUGCAUCUGUCUG---UGUGCGCUGCAUUUCAUCGCUAAUUUAAGUUAAUGUUCAUUGUUGUAAAUUACAUUAAUUUUAAUGCGAUUUCGCA ..--.....((((((..(((..(((......)---))..)))))))))..(((((......((((((((((..(((......))).))))))))))....)))))...... ( -26.10) >DroPse_CAF1 322811 93 + 1 CC--CUG---CACCAC---GCACCAUG----------UACAGUGCAUUUCAUCGCUAAUUUAAGUUAAUGGUCAUUGUUGUAAAUUACAUUAAUUUUAAUGCGAUUUCGCA ..--..(---(.....---((((....----------....)))).....(((((......(((((((((...(((......)))..)))))))))....)))))...)). ( -19.30) >DroGri_CAF1 342088 99 + 1 CGUCCUUUUGUUGA-------CCAUGUGUC-----CGUGCAUUGCAUUUCAUCGCUAAUUUAAGUUAAUGUUCAUUGUUGUAAAUUACAUUAAUUUUAAUGCGAUUUCGCA .(((........))-------)..((((..-----..((((((((........))......((((((((((..(((......))).)))))))))).))))))....)))) ( -16.70) >DroAna_CAF1 262220 101 + 1 AG--G-----AGUCCC---GCCUCCCUGGCCCCAACGAACAUUGCAUUUCAUCGCUAAUUUAAGUUAAUGGUCAUUGUUGUAAAUUACAUUAAUUUUAAUGCGAUUUCGCA .(--(-----....))---(((.....))).....((((.((((((((.............(((((((((...(((......)))..))))))))).)))))))))))).. ( -19.74) >DroPer_CAF1 285435 93 + 1 CC--CUG---CACCAC---GCACCAUG----------UACAGUGCAUUUCAUCGCUAAUUUAAGUUAAUGGUCAUUGUUGUAAAUUACAUUAAUUUUAAUGCGAUUUCGCA ..--..(---(.....---((((....----------....)))).....(((((......(((((((((...(((......)))..)))))))))....)))))...)). ( -19.30) >consensus CA__CUG___AACCAC___GCCCAUUUG_C_______UACACUGCAUUUCAUCGCUAAUUUAAGUUAAUGGUCAUUGUUGUAAAUUACAUUAAUUUUAAUGCGAUUUCGCA ..........................................(((.....(((((......(((((((((...(((......)))..)))))))))....)))))...))) (-12.60 = -12.60 + -0.00)
| Location | 15,490,294 – 15,490,395 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.02 |
| Mean single sequence MFE | -23.77 |
| Consensus MFE | -12.67 |
| Energy contribution | -13.33 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.755630 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
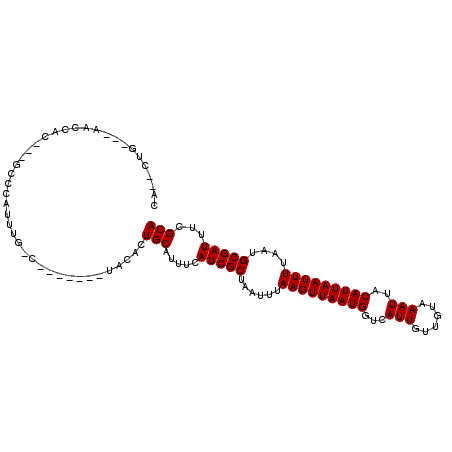
>2R_DroMel_CAF1 15490294 101 + 20766785 -AA---GCCACUGCAUUUCAUCGCUAAUUUAAGUUAAUGGUCAUUGUUGUAAAUUACAUUAAUUUUAAUGCGAUUUCGCACACACGCAUGCACCGG--AAGUGCUCC------------- -..---......((((((((((((......(((((((((...(((......)))..)))))))))....)))))...(((........)))....)--))))))...------------- ( -22.80) >DroVir_CAF1 381422 94 + 1 ----UGUGCGCUGCAUUUCAUCGCUAAUUUAAGUUAAUGUUCAUUGUUGUAAAUUACAUUAAUUUUAAUGCGAUUUCGCACACGCGCCUGCACUGGGC---------------------- ----...(((.(((.....(((((......((((((((((..(((......))).))))))))))....)))))...)))..)))((((.....))))---------------------- ( -22.90) >DroGri_CAF1 342111 116 + 1 ----CGUGCAUUGCAUUUCAUCGCUAAUUUAAGUUAAUGUUCAUUGUUGUAAAUUACAUUAAUUUUAAUGCGAUUUCGCACACGCGCCUGCACUGCGUAAGCGUCGGCACUGUUGUUGUA ----.((((..(((.....(((((......((((((((((..(((......))).))))))))))....)))))...)))..(((((.(((.....))).))).)))))).......... ( -28.60) >DroMoj_CAF1 434999 91 + 1 CUGCUGUGCGUUGCAUUUCAUCGCUAAUUUAAGUUAAUGUUCAUUGUUGUAAAUUACAUUAAUUUUAAUGCGAUUUCACACACACGCACAC----------------------------- ....(((((((((......(((((......((((((((((..(((......))).))))))))))....)))))......)).))))))).----------------------------- ( -23.00) >DroAna_CAF1 262242 104 + 1 -CAACGAACAUUGCAUUUCAUCGCUAAUUUAAGUUAAUGGUCAUUGUUGUAAAUUACAUUAAUUUUAAUGCGAUUUCGCACACACGCAUGCACCGG--AAGUGCCCC------------- -...........((((((((((((......(((((((((...(((......)))..)))))))))....)))))...(((........)))....)--))))))...------------- ( -22.40) >DroPer_CAF1 285454 99 + 1 ------UACAGUGCAUUUCAUCGCUAAUUUAAGUUAAUGGUCAUUGUUGUAAAUUACAUUAAUUUUAAUGCGAUUUCGCACACACGCAUGCACCGG--AAGUGCUCC------------- ------....(.((((((((((((......(((((((((...(((......)))..)))))))))....)))))...(((........)))....)--)))))).).------------- ( -22.90) >consensus _____GUGCAUUGCAUUUCAUCGCUAAUUUAAGUUAAUGGUCAUUGUUGUAAAUUACAUUAAUUUUAAUGCGAUUUCGCACACACGCAUGCACCGG__AAGUGC_CC_____________ ...........((((....(((((......(((((((((...(((......)))..)))))))))....)))))...((......)).))))............................ (-12.67 = -13.33 + 0.67)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:27:42 2006