
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 15,488,786 – 15,488,942 |
| Length | 156 |
| Max. P | 0.997490 |

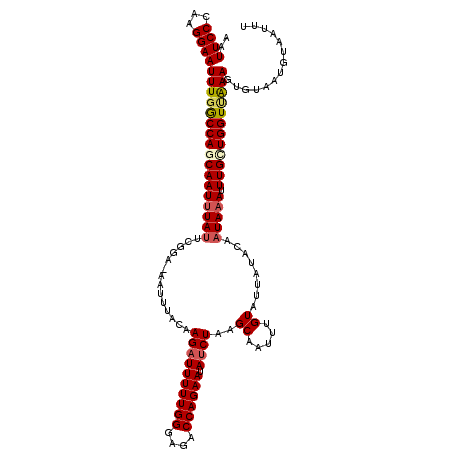
| Location | 15,488,786 – 15,488,906 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.88 |
| Mean single sequence MFE | -27.88 |
| Consensus MFE | -24.38 |
| Energy contribution | -25.06 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.87 |
| SVM decision value | 2.87 |
| SVM RNA-class probability | 0.997490 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
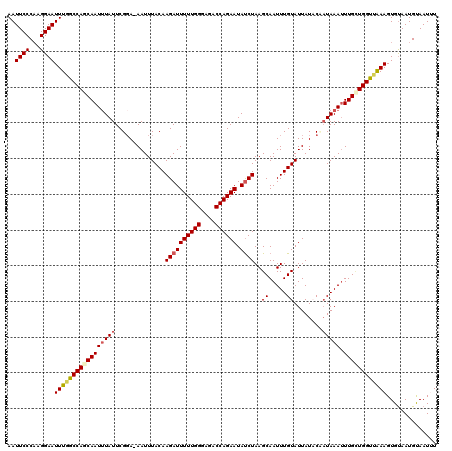
>2R_DroMel_CAF1 15488786 120 - 20766785 AAUUCCCAAGGAAUUUGGCCAGCAAUUUAUUCGCAAAAUUUACAAGAUUUUUGGGAGACCAGAAUAUCUAAGCAAUUUGUAUUAUCCUAUAAAUUUGCUGGUUAAAGUGCAAUGUAAUUU ..((((...))))(((((((((((....................((((((((((....)))))).))))....((((((((......))))))))))))))))))).............. ( -30.10) >DroSec_CAF1 281493 119 - 1 AAUUCCCAAGGAAUUUGGCCAGCAAUUUAUUCGGA-AAUUUACAAGAUUUUUGGGAGACCAGAAUAUCUAAGCAAUUUGUAUUAUACACUAAAUUUGCUGGUUAAAGUGUAAUGUAAUUU ..((((...))))((((((((((((((((......-........((((((((((....)))))).))))........((((...)))).)))).)))))))))))).............. ( -28.40) >DroSim_CAF1 260461 119 - 1 AAUUCCCAAGGAAUUUGGCCAACAAUUUAUUCGGA-AAUUUACAAGAUUUUUGGGAGACCAGAAUAUCUAAGCAAUUUGUAUUAUACAAUAAAUUUGCUGGUUAAAGUGUAAUGUAAUUU ..((((...))))(((((((...............-........((((((((((....)))))).)))).((((((((((........))))).)))))))))))).............. ( -24.70) >DroEre_CAF1 269682 119 - 1 AAUUCCCAAGGAAUUUGGCCAGCAAUGUAUUCGGA-AAUUUACAAGAUUUUUGGGAGACCAGAAUAUCUAAGCAAUUUGUAUUAUUCCAUAAAUUUGUUGGUAGAAGUGUAAUGCAAUGU ((((((...))))))..((((((((..(((..(((-(...((((((((((((((....)))))).))))........))))...)))))))...)))))))).................. ( -28.80) >DroYak_CAF1 279748 119 - 1 AAUUCCCCAGGAAUUUGACCAGCAAUUUAUUCGGA-AAUUUACAAGAUUUUUGGGAGACCAGAAUACCUAAGCAAUUUGUAUUAUACCAUAAACUUGGUGGUCAAAGUGUAAUGCAAUGU ..((((...))))((((((((.((((((((..((.-....((((((..((((((....))))))...........)))))).....))))))).))).)))))))).............. ( -27.42) >consensus AAUUCCCAAGGAAUUUGGCCAGCAAUUUAUUCGGA_AAUUUACAAGAUUUUUGGGAGACCAGAAUAUCUAAGCAAUUUGUAUUAUACAAUAAAUUUGCUGGUUAAAGUGUAAUGUAAUUU ..((((...))))(((((((((((((((((..............((((((((((....)))))).))))..((.....))........))))).)))))))))))).............. (-24.38 = -25.06 + 0.68)

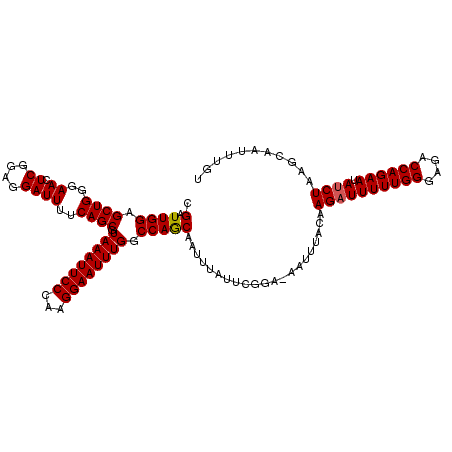
| Location | 15,488,826 – 15,488,942 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 97.57 |
| Mean single sequence MFE | -30.76 |
| Consensus MFE | -28.82 |
| Energy contribution | -28.86 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.58 |
| SVM RNA-class probability | 0.965178 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15488826 116 - 20766785 CGAUUGGAGCUGGGAACUCGGAGGAUUUUCAGCGUAAAUUCCCAAGGAAUUUGGCCAGCAAUUUAUUCGCAAAAUUUACAAGAUUUUUGGGAGACCAGAAUAUCUAAGCAAUUUGU .(.((((.((((..((.((....))))..)))).((((((((...)))))))).))))).........(((((.......((((((((((....)))))).))))......))))) ( -30.52) >DroSec_CAF1 281533 115 - 1 CGAUUGGAGCUGGGAACUCGGAGGAUUUUCAGCGUAAAUUCCCAAGGAAUUUGGCCAGCAAUUUAUUCGGA-AAUUUACAAGAUUUUUGGGAGACCAGAAUAUCUAAGCAAUUUGU .(((((..((((..((.((....))))..))))(((((((.((....((.(((.....))).))....)).-))))))).((((((((((....)))))).))))...)))))... ( -30.70) >DroSim_CAF1 260501 115 - 1 CGAUUGGAGCUGGGAACUCGGAGGAUUUUCAGCGUAAAUUCCCAAGGAAUUUGGCCAACAAUUUAUUCGGA-AAUUUACAAGAUUUUUGGGAGACCAGAAUAUCUAAGCAAUUUGU .(((((..((((..((.((....))))..))))(((((((.((....((.(((.....))).))....)).-))))))).((((((((((....)))))).))))...)))))... ( -31.70) >DroEre_CAF1 269722 115 - 1 CGAUUGGAGCUGGGAACUCGGAGGAUUUUCAGCGUAAAUUCCCAAGGAAUUUGGCCAGCAAUGUAUUCGGA-AAUUUACAAGAUUUUUGGGAGACCAGAAUAUCUAAGCAAUUUGU ....(((.((((..((.((....))))..)))).((((((((...)))))))).)))((..((((......-....))))((((((((((....)))))).))))..))....... ( -32.10) >DroYak_CAF1 279788 115 - 1 CGAUUGGAGCUGGGAACUCGGAGGAUUUUCAGCGUAAAUUCCCCAGGAAUUUGACCAGCAAUUUAUUCGGA-AAUUUACAAGAUUUUUGGGAGACCAGAAUACCUAAGCAAUUUGU ....(((.((((..((.((....))))..)))).((((((((...)))))))).)))((....((((((((-((((.....)))))))((....)).))))).....))....... ( -28.80) >consensus CGAUUGGAGCUGGGAACUCGGAGGAUUUUCAGCGUAAAUUCCCAAGGAAUUUGGCCAGCAAUUUAUUCGGA_AAUUUACAAGAUUUUUGGGAGACCAGAAUAUCUAAGCAAUUUGU .(.((((.((((..((.((....))))..)))).((((((((...)))))))).))))).....................((((((((((....)))))).))))........... (-28.82 = -28.86 + 0.04)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:27:40 2006