| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 15,456,596 – 15,456,690 |
| Length | 94 |
| Max. P | 0.745960 |

| Location | 15,456,596 – 15,456,690 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 76.12 |
| Mean single sequence MFE | -29.80 |
| Consensus MFE | -14.79 |
| Energy contribution | -15.15 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.745960 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
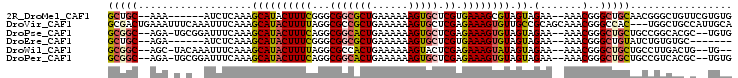
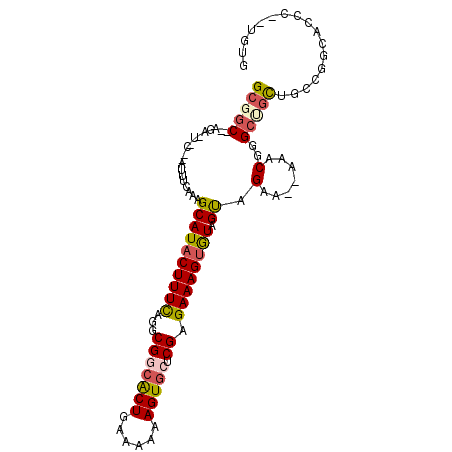
>2R_DroMel_CAF1 15456596 94 - 20766785 GCUGC--AAA------AUCUCAAAGCAUACUUUCGGGCGGCGCUGAAAAAAGUGCUCGUGAAAGCGUAGUAGAA--AAACGGGCUGCAACGGGCUGUUCGUGUG (((((--...------..((.((((....)))).))((((((((......))))).)))......)))))....--..((((((.((.....)).))))))... ( -25.70) >DroVir_CAF1 349553 101 - 1 GCGACUGAAAUUUCAAAUUUCAAAGCAUACUUUUAGGCGCCGCUGAAAAAAGUGCUCGAGAAAGUGUUGCCGCAGCAAACGGGCCAC---UGGCUGCCAUUGCA ((((.((((((.....))))))..((((((((((...((.((((......))))..)).))))))).))).(((((...(((....)---)))))))..)))). ( -27.20) >DroPse_CAF1 294605 97 - 1 GCGGC--AGA-UGCGGAUUUCAAAGCAUACUUUCAGGCGGCACUGAAAAAAGUGCUCGAGAAAGUGUAGUAGAA--AAACGGGCUGCUGCCGGCACGC--UGUG (((((--((.-(.((..((((...((((((((((...(((((((......))))).)).)))))))).)).)))--)..)).))))))))(((....)--)).. ( -37.10) >DroEre_CAF1 241651 87 - 1 GCUGC--AGA------AUCUCAAAGCAUACUUUCGGGCGGCGCUGAAAAAAGUGCUCGUGAAAGUGUAGUAGAA--AAACGGGCUGUAUCUGUGUGC------- (((((--(((------.......((((((((((((.(.((((((......))))))).))))))))).((....--..))..)))...)))))).))------- ( -26.50) >DroWil_CAF1 494962 95 - 1 GCGGC--AGC-UACAAAUUUCAAAGCAUACUUUUAGGCGCCACUGAAAAAAGUACUCGAGAAAGUAUAGUAGAA--AAACGGGCUGCUGCCUUGACUG--UG-- (((((--(((-(.....((((...((((((((((...((..(((......)))...)).)))))))).)).)))--)....)))))))))........--..-- ( -26.40) >DroPer_CAF1 256454 97 - 1 GCGGC--AGA-UGCGGAUUUCAAAGCAUACUUUCAGGCGGCACUGAAAAAAGUGCUCGAGAAAGUGUAGUAGAA--AAACGGGCUGCUGCCGUCACGC--UGUG (((((--((.-(.((..((((...((((((((((...(((((((......))))).)).)))))))).)).)))--)..)).))))))))........--.... ( -35.90) >consensus GCGGC__AGA_U_C__AUUUCAAAGCAUACUUUCAGGCGGCACUGAAAAAAGUGCUCGAGAAAGUGUAGUAGAA__AAACGGGCUGCUGCCGGCACCC__UGUG (((((...................((((((((((...(((((((......))))).)).)))))))).)).(.......)..)))))................. (-14.79 = -15.15 + 0.36)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:27:34 2006