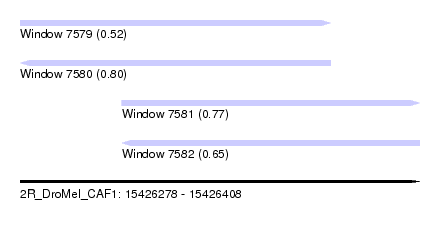
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 15,426,278 – 15,426,408 |
| Length | 130 |
| Max. P | 0.795530 |

| Location | 15,426,278 – 15,426,379 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 81.42 |
| Mean single sequence MFE | -31.20 |
| Consensus MFE | -20.91 |
| Energy contribution | -21.97 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.67 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.518207 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15426278 101 + 20766785 U-U-UAUAGAUAUAUGUAGAGAGUAGUGCGGAG-GCGG------CUGG--UCUGCGGCUAACAAAAGUUGUGCGCAGUCAGUUGACAUAAAUUUUGUUAGCCUACUUCCUAG .-.-............(((..(((((..((.((-((..------...)--))).))((((((((((.(((((..(((....))).))))).)))))))))))))))..))). ( -31.00) >DroPse_CAF1 270375 99 + 1 G-C------AGCUCUGGCGGCAGCGGUGCGGAGUGCGG------CUCGAUUCUGCGAGUAACAAAAUUUGUGCGCAGUCAGUUGACAUAAAUUUUGUUAGCCUACUUCCGGG (-(------(.(.(((....))).).)))((((((.((------((((......))).((((((((((((((..(((....))).))))))))))))))))))))))).... ( -33.30) >DroSim_CAF1 210370 102 + 1 U-UAUAUAGAUAUAUGUAGAGAGUAGUGCGGAG-GCGG------GUGG--UCUGCGGCUAACAAAAGUUGUGCGCAGUCAGUUGACAUAAAUUUUGUUAGCCUACUUCCCAG .-((((((....))))))(..(((((..((.((-((..------...)--))).))((((((((((.(((((..(((....))).))))).)))))))))))))))..)... ( -30.20) >DroEre_CAF1 218756 103 + 1 UUUACAUAGAUAUAUAUGGUGAGUGGUGCGGAG-GCGG------GUGG--CAUGCGGCUAACAAAAGUUGUGCGCAGUCAGUUGACAUAAAUUUUGUUAGCCUACUUCCUGG ....((((......))))(..((((..(((...-((..------...)--).)))(((((((((((.(((((..(((....))).))))).)))))))))))))))..)... ( -29.00) >DroYak_CAF1 226046 109 + 1 UUUAUAAAGAUAUAUAUAGAUAGUAGUGUGGAG-GCGGGUGCGGGUGG--UCUGCGGCUAACAAAAGUUGUGCGCAGUCAGUUGACAUAAAUUUUGUUAGCCUACUUCCUGG ((((((........)))))).........((((-(.....((((....--.))))(((((((((((.(((((..(((....))).))))).)))))))))))..)))))... ( -29.60) >DroPer_CAF1 231079 99 + 1 G-C------AGCUCUGUCGGCAGCGGUGCGGAGUGCGG------CUCGAUUCUGCGAGUAACAAAAUUUGUGCGCAGUCAGUUGACAUAAAUUUUGUUAGCCUACUUCCGGG (-(------(.(.(((....))).).)))((((((.((------((((......))).((((((((((((((..(((....))).))))))))))))))))))))))).... ( -34.10) >consensus U_U_UAUAGAUAUAUGUAGAGAGUAGUGCGGAG_GCGG______CUGG__UCUGCGGCUAACAAAAGUUGUGCGCAGUCAGUUGACAUAAAUUUUGUUAGCCUACUUCCUGG ................(((..((((..(((((...((........))...)))))(((((((((((.(((((..(((....))).))))).)))))))))))))))..))). (-20.91 = -21.97 + 1.06)

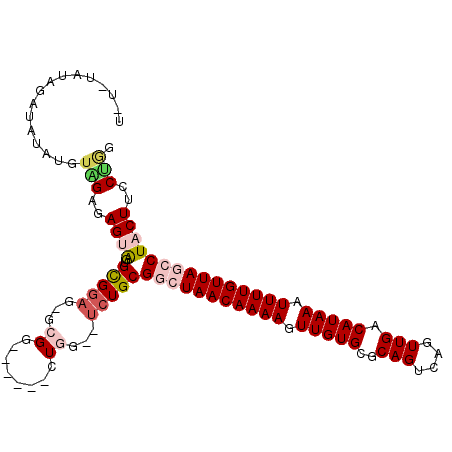

| Location | 15,426,278 – 15,426,379 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 81.42 |
| Mean single sequence MFE | -23.55 |
| Consensus MFE | -17.30 |
| Energy contribution | -17.13 |
| Covariance contribution | -0.17 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.795530 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15426278 101 - 20766785 CUAGGAAGUAGGCUAACAAAAUUUAUGUCAACUGACUGCGCACAACUUUUGUUAGCCGCAGA--CCAG------CCGC-CUCCGCACUACUCUCUACAUAUAUCUAUA-A-A .(((..(((((((((((((((.((.(((((......)).))).)).)))))))))))((.((--....------....-.)).))..))))..)))............-.-. ( -24.10) >DroPse_CAF1 270375 99 - 1 CCCGGAAGUAGGCUAACAAAAUUUAUGUCAACUGACUGCGCACAAAUUUUGUUACUCGCAGAAUCGAG------CCGCACUCCGCACCGCUGCCGCCAGAGCU------G-C ...(((.((.((((((((((((((.(((((......)).))).))))))))))).(((......))))------))..)))))(((.(.(((....))).).)------)-) ( -24.70) >DroSim_CAF1 210370 102 - 1 CUGGGAAGUAGGCUAACAAAAUUUAUGUCAACUGACUGCGCACAACUUUUGUUAGCCGCAGA--CCAC------CCGC-CUCCGCACUACUCUCUACAUAUAUCUAUAUA-A .(((..(((((((((((((((.((.(((((......)).))).)).)))))))))))((.((--....------....-.)).))..))))..)))..............-. ( -23.80) >DroEre_CAF1 218756 103 - 1 CCAGGAAGUAGGCUAACAAAAUUUAUGUCAACUGACUGCGCACAACUUUUGUUAGCCGCAUG--CCAC------CCGC-CUCCGCACCACUCACCAUAUAUAUCUAUGUAAA ...((.....(((((((((((.((.(((((......)).))).)).))))))))))).....--))..------..((-....)).........((((......)))).... ( -20.90) >DroYak_CAF1 226046 109 - 1 CCAGGAAGUAGGCUAACAAAAUUUAUGUCAACUGACUGCGCACAACUUUUGUUAGCCGCAGA--CCACCCGCACCCGC-CUCCACACUACUAUCUAUAUAUAUCUUUAUAAA ...(((.((.(((((((((((.((.(((((......)).))).)).)))))))))))((.(.--....).))....))-.)))............................. ( -22.70) >DroPer_CAF1 231079 99 - 1 CCCGGAAGUAGGCUAACAAAAUUUAUGUCAACUGACUGCGCACAAAUUUUGUUACUCGCAGAAUCGAG------CCGCACUCCGCACCGCUGCCGACAGAGCU------G-C ...(((.((.((((((((((((((.(((((......)).))).))))))))))).(((......))))------))..)))))(((.(.(((....))).).)------)-) ( -25.10) >consensus CCAGGAAGUAGGCUAACAAAAUUUAUGUCAACUGACUGCGCACAACUUUUGUUAGCCGCAGA__CCAC______CCGC_CUCCGCACCACUCUCUACAUAUAUCUAUA_A_A ..(((.(((.(((((((((((.((.(((((......)).))).)).)))))))))))...................((.....))...))).)))................. (-17.30 = -17.13 + -0.17)

| Location | 15,426,311 – 15,426,408 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 80.14 |
| Mean single sequence MFE | -31.82 |
| Consensus MFE | -17.49 |
| Energy contribution | -17.68 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.765745 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15426311 97 + 20766785 G-G------CUGG--UCUGCGGCUAACAAAAGUUGUGCGCAGUCAGUUGACAUAAAUUUUGUUAGCCUACUUCCUAGCGAAAUUGCUUACAGGCAAUGCGCAG-------GCA---- .-.------...(--((((((((((((((((.(((((..(((....))).))))).))))))))))).........((...((((((....))))))))))))-------)).---- ( -36.00) >DroPse_CAF1 270404 99 + 1 G-G------CUCGAUUCUGCGAGUAACAAAAUUUGUGCGCAGUCAGUUGACAUAAAUUUUGUUAGCCUACUUCCGGGCCAAAUUGCUUACAGACAAUGCACAG-------GCG---- (-(------((((...(((((....(((.....))).)))))...(((((((.......))))))).......)))))).....((((.((.....))...))-------)).---- ( -27.00) >DroGri_CAF1 277819 106 + 1 GGGC-------UGCGAGUGUGGCUAACAAAAGUUGUGCGCAGUCAGUUGACAUAAAUUUUGUUAGCCUACUUUUGGCCAAAAUUGCUUACAGACAAUGCACAGAAGCAAUGCG---- .(((-------((.(((((.(((((((((((.(((((..(((....))).))))).)))))))))))))))).)))))...(((((((.((.....)).....)))))))...---- ( -36.00) >DroWil_CAF1 454651 94 + 1 G-----------G--GUUGUGGCUAACAAAAGUUGUGCGCAGUCAGUUGACAUAAAUUUUGUUAGCCUACUUUUAGGAGAAAUUGCUUAC---CAUGGCCCAU-------GCAAGAG (-----------(--((..((((((((((((.(((((..(((....))).))))).)))))))))((((....))))............)---))..))))..-------....... ( -29.10) >DroYak_CAF1 226081 103 + 1 G-GGUGCGGGUGG--UCUGCGGCUAACAAAAGUUGUGCGCAGUCAGUUGACAUAAAUUUUGUUAGCCUACUUCCUGGCGAAAUUGCUUACAGGCAAUGCGCAG-------GCA---- .-..........(--((((((((((((((((.(((((..(((....))).))))).))))))))))).........((...((((((....))))))))))))-------)).---- ( -35.80) >DroPer_CAF1 231108 99 + 1 G-G------CUCGAUUCUGCGAGUAACAAAAUUUGUGCGCAGUCAGUUGACAUAAAUUUUGUUAGCCUACUUCCGGGCCAAAUUGCUUACAGACAAUGCACAG-------GCG---- (-(------((((...(((((....(((.....))).)))))...(((((((.......))))))).......)))))).....((((.((.....))...))-------)).---- ( -27.00) >consensus G_G______CUCG__UCUGCGGCUAACAAAAGUUGUGCGCAGUCAGUUGACAUAAAUUUUGUUAGCCUACUUCCGGGCGAAAUUGCUUACAGACAAUGCACAG_______GCA____ .................((((((((((((((.(((((..(((....))).))))).)))))))))))................(((...........)))..........))).... (-17.49 = -17.68 + 0.20)
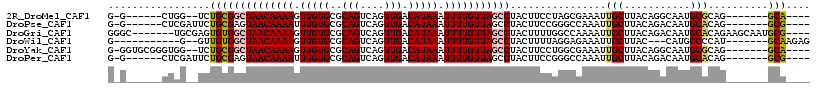
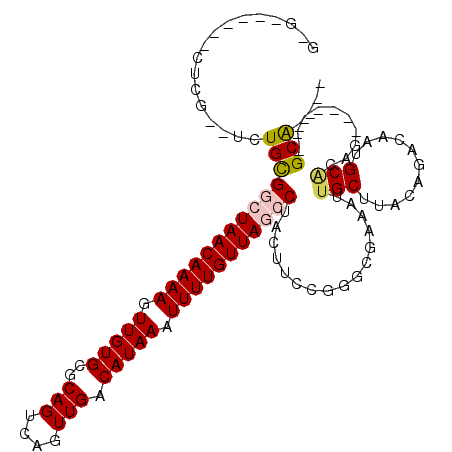

| Location | 15,426,311 – 15,426,408 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 80.14 |
| Mean single sequence MFE | -25.43 |
| Consensus MFE | -17.45 |
| Energy contribution | -18.12 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.648784 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15426311 97 - 20766785 ----UGC-------CUGCGCAUUGCCUGUAAGCAAUUUCGCUAGGAAGUAGGCUAACAAAAUUUAUGUCAACUGACUGCGCACAACUUUUGUUAGCCGCAGA--CCAG------C-C ----..(-------(((((.(((((......)))))..))).))).....(((((((((((.((.(((((......)).))).)).)))))))))))((...--...)------)-. ( -28.40) >DroPse_CAF1 270404 99 - 1 ----CGC-------CUGUGCAUUGUCUGUAAGCAAUUUGGCCCGGAAGUAGGCUAACAAAAUUUAUGUCAACUGACUGCGCACAAAUUUUGUUACUCGCAGAAUCGAG------C-C ----...-------....((.(((((((..((......(((((....)..))))((((((((((.(((((......)).))).)))))))))).))..))))..))))------)-. ( -23.40) >DroGri_CAF1 277819 106 - 1 ----CGCAUUGCUUCUGUGCAUUGUCUGUAAGCAAUUUUGGCCAAAAGUAGGCUAACAAAAUUUAUGUCAACUGACUGCGCACAACUUUUGUUAGCCACACUCGCA-------GCCC ----.((..(((((.((.(((((((......)))))....)))).)))))(((((((((((.((.(((((......)).))).)).)))))))))))......)).-------.... ( -27.30) >DroWil_CAF1 454651 94 - 1 CUCUUGC-------AUGGGCCAUG---GUAAGCAAUUUCUCCUAAAAGUAGGCUAACAAAAUUUAUGUCAACUGACUGCGCACAACUUUUGUUAGCCACAAC--C-----------C ...((((-------....((....---))..))))............((.(((((((((((.((.(((((......)).))).)).)))))))))))))...--.-----------. ( -21.20) >DroYak_CAF1 226081 103 - 1 ----UGC-------CUGCGCAUUGCCUGUAAGCAAUUUCGCCAGGAAGUAGGCUAACAAAAUUUAUGUCAACUGACUGCGCACAACUUUUGUUAGCCGCAGA--CCACCCGCACC-C ----..(-------(((.(((((((......)))))...)))))).....(((((((((((.((.(((((......)).))).)).)))))))))))((.(.--....).))...-. ( -28.90) >DroPer_CAF1 231108 99 - 1 ----CGC-------CUGUGCAUUGUCUGUAAGCAAUUUGGCCCGGAAGUAGGCUAACAAAAUUUAUGUCAACUGACUGCGCACAAAUUUUGUUACUCGCAGAAUCGAG------C-C ----...-------....((.(((((((..((......(((((....)..))))((((((((((.(((((......)).))).)))))))))).))..))))..))))------)-. ( -23.40) >consensus ____CGC_______CUGUGCAUUGUCUGUAAGCAAUUUCGCCCGGAAGUAGGCUAACAAAAUUUAUGUCAACUGACUGCGCACAACUUUUGUUAGCCGCAGA__CCAG______C_C ..............((((..(((((......)))))..............(((((((((((.((.(((((......)).))).)).)))))))))))))))................ (-17.45 = -18.12 + 0.67)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:27:25 2006