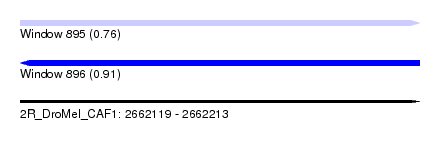
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 2,662,119 – 2,662,213 |
| Length | 94 |
| Max. P | 0.907390 |

| Location | 2,662,119 – 2,662,213 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 89.53 |
| Mean single sequence MFE | -31.85 |
| Consensus MFE | -21.53 |
| Energy contribution | -21.66 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.85 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.755631 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
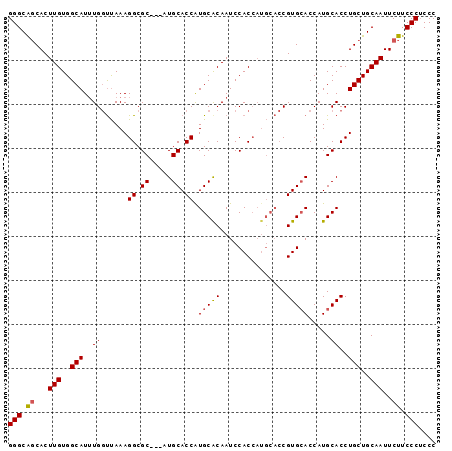
>2R_DroMel_CAF1 2662119 94 + 20766785 GGGCAUCACUUGUGGCAUUUGGUUAAAGGCGC---AUGCACCGUGCACAAUCCACCAUGCACCGUGGACCAUGCACCUGCUGCAAUUCUUCCCUCCC (((......(((..(((..(((((...((.((---(((....(((.......)))))))).))...)))))......)))..))).....))).... ( -28.20) >DroSec_CAF1 95370 97 + 1 GGGCAGCACUUGUGGCAUUUGGUUAAAGGCGCACCAUGCACCAUGCACAAUCCACCAUGCACCGUGCACCAUGCACCUGCUGCAAUUCUUCCCUCCC (((.((...(((..(((..........((.(((...(((((..((((..........))))..)))))...))).)))))..)))..)).))).... ( -28.10) >DroSim_CAF1 93444 94 + 1 GGGCAGCACUUGUGGCAUAUGGUUAAAGGCGC---AUGCACCAUGCGCAAUCCACCAUGCACCGUGCACCAUGCACCUGCUGCAAUUCUUCCCUCCC (((.((...(((..(((((((((.....((((---(((...))))))).....))))))....((((.....)))).)))..)))..)).))).... ( -36.00) >DroEre_CAF1 94002 87 + 1 GGGCGGCACUUGUGGCAUUUGCCCAAAGGCGC---AUGCACCAUGCACAAUCCAC-------CGUGCACCAUGCACCUGCUGCAAUUCUUCCCGCCC ((((((...(((..(((...(((....)))((---(((.....(((((.......-------.))))).)))))...)))..)))......)))))) ( -35.10) >consensus GGGCAGCACUUGUGGCAUUUGGUUAAAGGCGC___AUGCACCAUGCACAAUCCACCAUGCACCGUGCACCAUGCACCUGCUGCAAUUCUUCCCUCCC (((.((...(((..(((..((......((.((.....)).)).(((((...............))))).....))..)))..)))..)).))).... (-21.53 = -21.66 + 0.13)

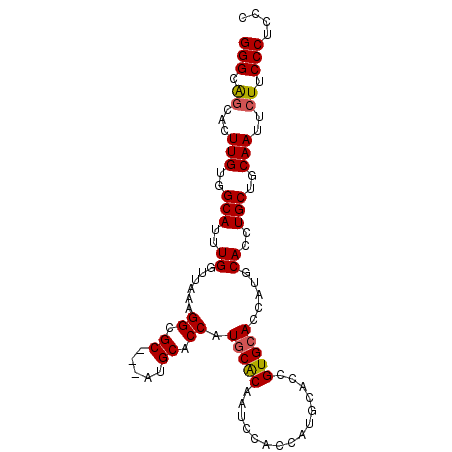
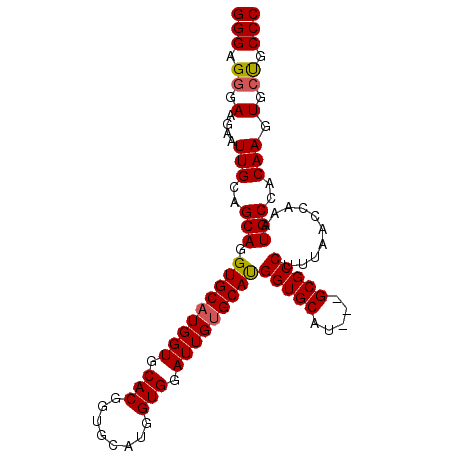
| Location | 2,662,119 – 2,662,213 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 89.53 |
| Mean single sequence MFE | -39.40 |
| Consensus MFE | -28.95 |
| Energy contribution | -29.07 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.78 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.907390 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2662119 94 - 20766785 GGGAGGGAAGAAUUGCAGCAGGUGCAUGGUCCACGGUGCAUGGUGGAUUGUGCACGGUGCAU---GCGCCUUUAACCAAAUGCCACAAGUGAUGCCC ....(((.....(((..(((.(((((..((((((........))))))..)))))((((...---.))))..........)))..)))......))) ( -35.60) >DroSec_CAF1 95370 97 - 1 GGGAGGGAAGAAUUGCAGCAGGUGCAUGGUGCACGGUGCAUGGUGGAUUGUGCAUGGUGCAUGGUGCGCCUUUAACCAAAUGCCACAAGUGCUGCCC ..............(((((((((((((.((((((.(((((..(....)..))))).)))))).))))))))..........((.....))))))).. ( -42.10) >DroSim_CAF1 93444 94 - 1 GGGAGGGAAGAAUUGCAGCAGGUGCAUGGUGCACGGUGCAUGGUGGAUUGCGCAUGGUGCAU---GCGCCUUUAACCAUAUGCCACAAGUGCUGCCC ..............((((((.((((.....))))(((..(((((.((..(((((((...)))---))))..)).)))))..))).....)))))).. ( -38.10) >DroEre_CAF1 94002 87 - 1 GGGCGGGAAGAAUUGCAGCAGGUGCAUGGUGCACG-------GUGGAUUGUGCAUGGUGCAU---GCGCCUUUGGGCAAAUGCCACAAGUGCCGCCC ((((((.(....(((..(((.((((((.(((((((-------(....)))))))).))))))---..(((....)))...)))..))).).)))))) ( -41.80) >consensus GGGAGGGAAGAAUUGCAGCAGGUGCAUGGUGCACGGUGCAUGGUGGAUUGUGCAUGGUGCAU___GCGCCUUUAACCAAAUGCCACAAGUGCUGCCC (((.((.(....(((..(((.(((((((((.(((........))).)))))))))(((((.....)))))..........)))..))).).)).))) (-28.95 = -29.07 + 0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:39:25 2006