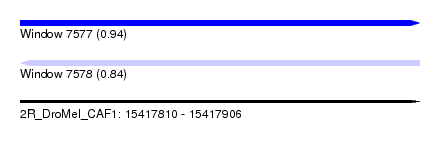
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 15,417,810 – 15,417,906 |
| Length | 96 |
| Max. P | 0.938091 |

| Location | 15,417,810 – 15,417,906 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 78.87 |
| Mean single sequence MFE | -27.53 |
| Consensus MFE | -12.94 |
| Energy contribution | -13.30 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.82 |
| Structure conservation index | 0.47 |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.938091 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15417810 96 + 20766785 AACGAUCGUUUUGUCAUAAUGCGACAUUUAAGGAAUUUUUAUAUGAAUUCACUUAACGACUUG--------GCAUUGGAACGAUUCGCGACGAUCCUGUCGCGU ...(((((((((((((.....((....(((((((((((......)))))).)))))))...))--------)))...))))))))(((((((....))))))). ( -28.10) >DroVir_CAF1 302824 100 + 1 AACGAUCGUUUUGUCAUAAUGCGACAUUUAAGGAAUUUUUAUAUGAAUUCACUUAACGAUCUGACGAUCUGGCAUUAG--CGA--CGCGUCGCUCCUGGCGCGU ...(((((((.((((.......)))).(((((((((((......)))))).)))))......)))))))..((....)--).(--(((((((....)))))))) ( -29.30) >DroPse_CAF1 261135 91 + 1 AACGAUCGUUUUGUCAUAAUGCGACAUUUAAGGAAUUUUUAUAUGAAUUCACUUAACGACUUC--------GCAUUGGGAC-----GCGACGAUCCUGGCGCGU ...(((((((.((((.((((((((...(((((((((((......)))))).))))).....))--------)))))).)))-----).)))))))......... ( -31.40) >DroGri_CAF1 266114 92 + 1 AGCGAUCGUUUUGUCAUAAUGCGACAUUUAAGGAAUUUUUAUAUGAAUUCACUUAACGAUCUCUGAAUC-GACGUUGC--UGA--CGCUUCGAUCCU------- ...(((((...(((((....(((((..(((((((((((......)))))).)))))((((......)))-)..)))))--)))--))...)))))..------- ( -26.30) >DroMoj_CAF1 336454 89 + 1 AACGAUCGUUUUGUCAUAAUGCGACAUUUAAGGAAUCUCUAUAUGAAUUCACUUAACGAUCUGACGAUCUG-----------A--CGCGUUGCUCCUC--GGUU ...(((((....((...((((((....(((((((((.((.....)))))).))))).((((....))))..-----------.--))))))))....)--)))) ( -18.70) >DroPer_CAF1 221875 91 + 1 AACGAUCGUUUUGUCAUAAUGCGACAUUUAAGGAAUUUUUAUAUGAAUUCACUUAACGACUUC--------GCAUUGGGAC-----GCGACGAUCCUGGCGCGU ...(((((((.((((.((((((((...(((((((((((......)))))).))))).....))--------)))))).)))-----).)))))))......... ( -31.40) >consensus AACGAUCGUUUUGUCAUAAUGCGACAUUUAAGGAAUUUUUAUAUGAAUUCACUUAACGACCUC________GCAUUGG__C_A__CGCGACGAUCCUGGCGCGU ...(((((...((((.......)))).(((((((((((......)))))).)))))..................................)))))......... (-12.94 = -13.30 + 0.36)

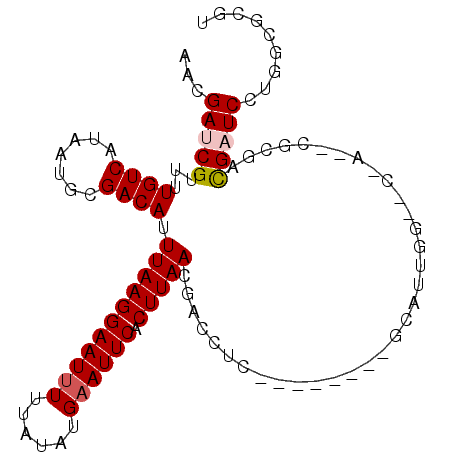
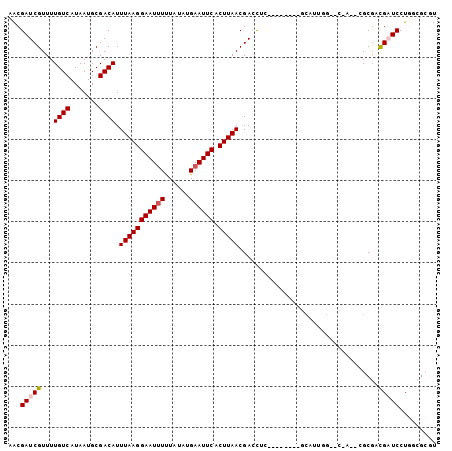
| Location | 15,417,810 – 15,417,906 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 78.87 |
| Mean single sequence MFE | -24.18 |
| Consensus MFE | -12.84 |
| Energy contribution | -13.20 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.843152 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15417810 96 - 20766785 ACGCGACAGGAUCGUCGCGAAUCGUUCCAAUGC--------CAAGUCGUUAAGUGAAUUCAUAUAAAAAUUCCUUAAAUGUCGCAUUAUGACAAAACGAUCGUU .((((((......)))))).((((((.......--------.......(((((.(((((........)))))))))).(((((.....))))).)))))).... ( -23.00) >DroVir_CAF1 302824 100 - 1 ACGCGCCAGGAGCGACGCG--UCG--CUAAUGCCAGAUCGUCAGAUCGUUAAGUGAAUUCAUAUAAAAAUUCCUUAAAUGUCGCAUUAUGACAAAACGAUCGUU ....(.((..(((((....--)))--))..)).).((((((.......(((((.(((((........)))))))))).(((((.....)))))..))))))... ( -25.40) >DroPse_CAF1 261135 91 - 1 ACGCGCCAGGAUCGUCGC-----GUCCCAAUGC--------GAAGUCGUUAAGUGAAUUCAUAUAAAAAUUCCUUAAAUGUCGCAUUAUGACAAAACGAUCGUU .........((((((...-----(((..(((((--------((.....(((((.(((((........))))))))))...)))))))..)))...))))))... ( -26.40) >DroGri_CAF1 266114 92 - 1 -------AGGAUCGAAGCG--UCA--GCAACGUC-GAUUCAGAGAUCGUUAAGUGAAUUCAUAUAAAAAUUCCUUAAAUGUCGCAUUAUGACAAAACGAUCGCU -------.(((((((.((.--...--))....))-)))))((.((((((((((.(((((........))))))))...(((((.....))))).))))))).)) ( -25.40) >DroMoj_CAF1 336454 89 - 1 AACC--GAGGAGCAACGCG--U-----------CAGAUCGUCAGAUCGUUAAGUGAAUUCAUAUAGAGAUUCCUUAAAUGUCGCAUUAUGACAAAACGAUCGUU ...(--((.((((...)).--)-----------)...)))...((((((((((.(((((........))))))))...(((((.....))))).)))))))... ( -18.50) >DroPer_CAF1 221875 91 - 1 ACGCGCCAGGAUCGUCGC-----GUCCCAAUGC--------GAAGUCGUUAAGUGAAUUCAUAUAAAAAUUCCUUAAAUGUCGCAUUAUGACAAAACGAUCGUU .........((((((...-----(((..(((((--------((.....(((((.(((((........))))))))))...)))))))..)))...))))))... ( -26.40) >consensus ACGCGCCAGGAUCGACGCG__U_G__CCAAUGC________CAAAUCGUUAAGUGAAUUCAUAUAAAAAUUCCUUAAAUGUCGCAUUAUGACAAAACGAUCGUU .........(((((..................................(((((.(((((........)))))))))).(((((.....)))))...)))))... (-12.84 = -13.20 + 0.36)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:27:21 2006