
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 15,411,990 – 15,412,090 |
| Length | 100 |
| Max. P | 0.603054 |

| Location | 15,411,990 – 15,412,090 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 76.66 |
| Mean single sequence MFE | -29.67 |
| Consensus MFE | -13.82 |
| Energy contribution | -12.80 |
| Covariance contribution | -1.02 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.47 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.603054 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15411990 100 + 20766785 AGCA-AUAAUCAGUGUGC--UGCUGUGGCGACCA-UAAAACGCCAAA-UGCUGCGUAAUGUAUGCAACAUAAAUGCA--GAUAUUUGCGGCACGGCCGGCUAACUGG ....-.....((((..((--((((((((((....-.....))))...-.((((((.(((((.((((.......))))--.))))))))))))))).))))..)))). ( -35.10) >DroVir_CAF1 292995 92 + 1 AGCAAAUAAUCAGAGU--GUUGUUGUGGCAACAACCAAAACGCCAAA-UGUUGCGUAAUGUAUGCAACAUAGCUGCA--GAUAGUCGCGGCCGUGCA---------- .(((........(.((--((((((((....)))))...))))))..(-(((((((((...)))))))))).(((((.--.......)))))..))).---------- ( -28.20) >DroPse_CAF1 247462 94 + 1 AGCA-AUAAUCAGAGCAC--AGCUGCGGCCGCUU-UGAAACGCCAAA-UUCUGCGUAAUGUAUGCAACAUGGCUGGA--GAUAGUUGCGGCCUGG------AAAUGG .(((-....(((((((..--.((....)).))))-))).((((....-....))))......)))..((.(((((.(--(....)).))))))).------...... ( -25.20) >DroWil_CAF1 429475 96 + 1 AGCA-AUAAUCAGUGUGU--UGCUGUGGCCACCA-UGAAACGCCAAA-UUUUGCCUAAUGCAUGCAACAUAACUGCAUAGAUAGUUGCGGUCUGCU-G---AAGU-- ....-....(((((((((--(((..((((.....-......))))..-...(((.....))).))))))).((((((........))))))..)))-)---)...-- ( -25.80) >DroYak_CAF1 211431 101 + 1 AGCA-AUAAUCAGUGUGC--UGCUGUGGCGACUA-UAAAACGCCAAAGUGCUGCGUAAUGUAUGCAACAUAAAUGCA--GAUAUUUGCGGCACGGCCGGCGAACUGG ....-.....((((.(((--((((.(((((....-.....)))))..((((((((.(((((.((((.......))))--.))))))))))))))).))))).)))). ( -38.50) >DroPer_CAF1 207455 94 + 1 AGCA-AUAAUCAGAGCAC--AGCUGCGGCCGCUU-UGAAACGCCAAA-UUCUGCGUAAUGUAUGCAACAUGGCUGGA--GAUAGUUGCGGCCUGG------AAAUGG .(((-....(((((((..--.((....)).))))-))).((((....-....))))......)))..((.(((((.(--(....)).))))))).------...... ( -25.20) >consensus AGCA_AUAAUCAGAGUGC__UGCUGUGGCCACUA_UAAAACGCCAAA_UGCUGCGUAAUGUAUGCAACAUAACUGCA__GAUAGUUGCGGCCUGGC_____AAAUGG .....................((((((((............))).......(((((.....)))))....(((((......))))))))))................ (-13.82 = -12.80 + -1.02)

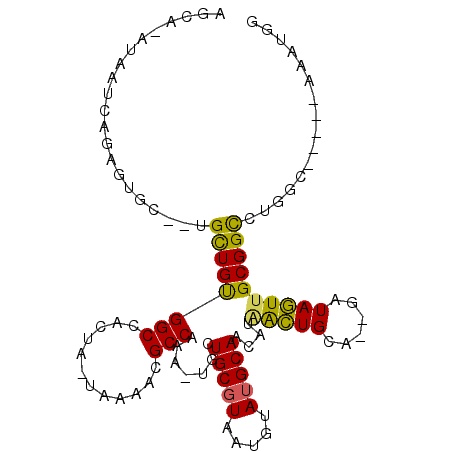
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:27:19 2006