| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 15,407,406 – 15,407,496 |
| Length | 90 |
| Max. P | 0.534746 |

| Location | 15,407,406 – 15,407,496 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 83.18 |
| Mean single sequence MFE | -38.22 |
| Consensus MFE | -24.45 |
| Energy contribution | -24.23 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.534746 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
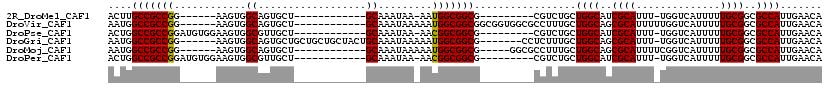
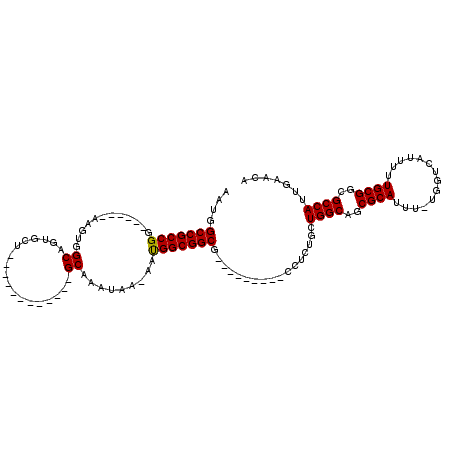
>2R_DroMel_CAF1 15407406 90 + 20766785 ACUUGCCGCCGG------AAGUGGCAGUGCU------------GCAAAUAA-AAUGGCGGCG---------CGUCUGCUGGCAUCGCAUUU-UGGUCAUUUUUGCGGCGCCAUUGAACA ...(((((.(..------..))))))(((((------------((......-....))))))---------)((..(.((((..((((...-..........))))..)))).)..)). ( -33.32) >DroVir_CAF1 285794 101 + 1 AAUGGCCGCCGG------AAGUGGCAGUGCU------------GCAAAUAAAAAUGGCGGCGGCGGUGGCGCCUUUGCUGGCAGCGCAUUUUUGGUCAUUUUUGCGGCGCCAUUGAACA ((((((.(((((------(((((((.(((((------------((..........(((((.((((....)))).))))).))))))).......))))))))..))))))))))..... ( -42.40) >DroPse_CAF1 242421 96 + 1 ACUGGCCGCCGGAUGUGGAAGUGGCGUUGCU------------GCAAAUAA-AACGGCGGCG---------CGUCUGCUGGCAUCGCAUUU-UGGUCAUUUUUGCGGCGCCAUUGAACA ..((((.(((((((((.((((((((((((((------------(.......-..))))))))---------)(((....)))....)))))-).).))))....))))))))....... ( -35.30) >DroGri_CAF1 250242 105 + 1 AAUGGCCGCCGG------AAGUGGCAGUGCUGCUGCUGCUACUGCAAAUAAAAAUGGCGGCG-------CCUCUUUGCUGGCAGCGCAUUU-UGGUCAUUUUUGCGGCGCCAUUGAACA ((((((.(((((------(((((((.(((((((((((((((.............)))))))(-------(......)).))))))))....-..))))))))..))))))))))..... ( -43.62) >DroMoj_CAF1 309795 96 + 1 AAUGGCCGCCGG------AAGUGGCAGUGCU------------GCAAAUAAAAAUGGCGGCG-----GGCGCCUUUGCUGGCAGCGCAUUUUCGGUCAUUUUUGCGGCGCCAUUGAACA ((((((.(((((------(((((((.(((((------------((......(((.((((...-----..)))))))....))))))).......))))))))..))))))))))..... ( -39.40) >DroPer_CAF1 202460 96 + 1 ACUGGCCGCCGGAUGUGGAAGUGGCGUUGCU------------GCAAAUAA-AACGGCGGCG---------CGUCUGCUGGCAUCGCAUUU-UGGUCAUUUUUGCGGCGCCAUUGAACA ..((((.(((((((((.((((((((((((((------------(.......-..))))))))---------)(((....)))....)))))-).).))))....))))))))....... ( -35.30) >consensus AAUGGCCGCCGG______AAGUGGCAGUGCU____________GCAAAUAA_AAUGGCGGCG_________CCUCUGCUGGCAGCGCAUUU_UGGUCAUUUUUGCGGCGCCAUUGAACA ....(((((((............((..................)).........))))))).................((((..((((..............))))..))))....... (-24.45 = -24.23 + -0.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:27:16 2006