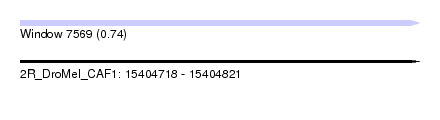
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 15,404,718 – 15,404,821 |
| Length | 103 |
| Max. P | 0.740836 |

| Location | 15,404,718 – 15,404,821 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 84.43 |
| Mean single sequence MFE | -23.77 |
| Consensus MFE | -18.24 |
| Energy contribution | -17.93 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.740836 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
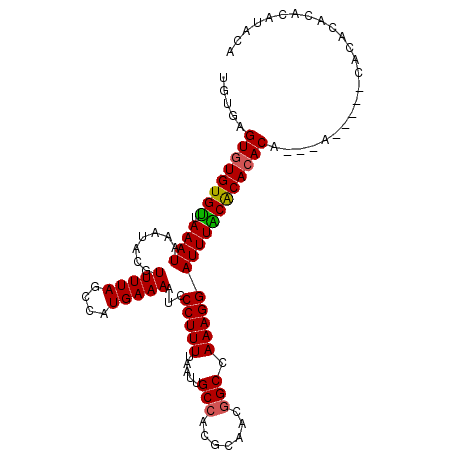
>2R_DroMel_CAF1 15404718 103 + 20766785 UGUGAGUGUGUGUUAAAUAAAUACGUUUUAGCCAUGAAAAUCCCUUUUAAUUGCCACGCAACGGCCAAAGGAUUUGCGCACAC----ACACACCACACUCACGUACA .((((((((((((....((((.....))))((.....((((((.(((.....(((.......))).)))))))))..))....----..))).)))))))))..... ( -24.60) >DroPse_CAF1 239564 100 + 1 UAUAUGUGUGUGUUAAAUAAAUACGUUUUAGUCAUGAAAAUCCCUUUUAAUUGCCACGCAACGGCAAAAGGAUUUACACACACAAG-------CAGACAGAGAUACA ....(((((((((.((((.......(((((....)))))...(((((....((((.......))))))))))))))))))))))..-------.............. ( -22.70) >DroEre_CAF1 197291 105 + 1 UGUGAGUGUGUGCUAAAUAAAUACGUUUUAGCCAUGAAAAUCCCUUUUAAUUGCCACGCAACGGCCAAAGGAUUUGCGCACACA--CACUCGCCACACACACGUACA .((((((((((((((((.........))))))..((.((((((.(((.....(((.......))).)))))))))...)).)))--))))))).............. ( -27.50) >DroYak_CAF1 204113 105 + 1 UGUGAGUGUGUGUUAAAUAAAUACGUUUUAGCCAUGAAAAUCCCUUUUAAUUGCCACGCAACGGCCAAAGGAUUUGCGCACACA--CACUCACCACACACAUGUACA .((((((((((((....(((((...((((.(((.(((((.....))))).((((...)))).))).)))).)))))...)))))--))))))).............. ( -27.50) >DroAna_CAF1 187740 96 + 1 GGAGUGUCUGUGUUAAAUAAAUACGUUUUAGCCAUGAAAAUCCCUUUUAAUUGCCACGCAACAGCCAAAGGAUUUACACACACA--CA---------CGAAUAUUAA ...((((.((((((((((.......(((((....)))))...(((((...((((...)))).....)))))))))).))))).)--))---------)......... ( -17.30) >DroPer_CAF1 199587 100 + 1 UAUGUGUGUGUGUUAAAUAAAUACGUUUUAGUCAUGAAAAUCCCUUUUAAUUGCCACGCAACGGCAAAAGGAUUUACACACACAAG-------CAGACAGAGAUACA ..((((((((((.............(((((....)))))...(((((....((((.......)))))))))...))))))))))..-------.............. ( -23.00) >consensus UGUGAGUGUGUGUUAAAUAAAUACGUUUUAGCCAUGAAAAUCCCUUUUAAUUGCCACGCAACGGCCAAAGGAUUUACACACACA___A_____CACACACACAUACA .....((((((((.((((.......(((((....)))))...(((((.....(((.......))).)))))))))))))))))........................ (-18.24 = -17.93 + -0.30)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:27:13 2006