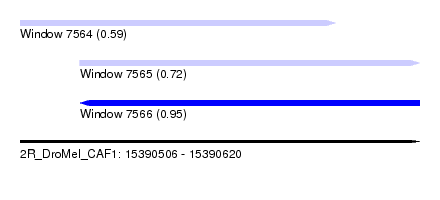
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 15,390,506 – 15,390,620 |
| Length | 114 |
| Max. P | 0.953794 |

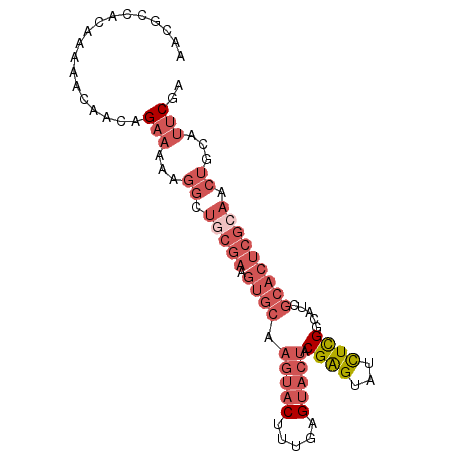
| Location | 15,390,506 – 15,390,596 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 90 |
| Reading direction | forward |
| Mean pairwise identity | 82.96 |
| Mean single sequence MFE | -23.73 |
| Consensus MFE | -13.81 |
| Energy contribution | -16.32 |
| Covariance contribution | 2.50 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.593779 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
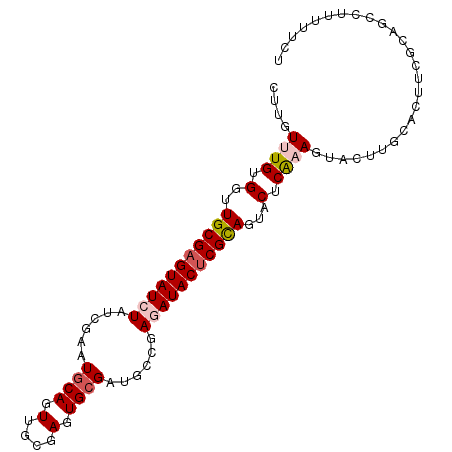
>2R_DroMel_CAF1 15390506 90 + 20766785 AACGCCACAAAAACAACAGAAAAAGGCUGCGAAGUGCAAGUACUUUGAGUACUACGAGUAUCUCGGCAUCGCACUCGCAACUGCAUUCGA ..................(((...((.(((((.((((.(((((.....))))).((((...)))).....))))))))).))...))).. ( -25.00) >DroPse_CAF1 225044 69 + 1 UCCACCG-AAAAACUGCAGCCA-----------------CAACUGCGAG---UGCGAGUAUCUUGUCCUCGCACUCGCAACUGCAUCCGA .......-......(((((...-----------------....((((((---((((((.........)))))))))))).)))))..... ( -25.50) >DroSec_CAF1 196083 90 + 1 AACGCCACAAAAACAACAGAAAAAGGCUGCGAAGUGCAAGUACUUUGAGUACUACGAGUAUCUCGGCAUCGCACUCGCAACUGAAUUCGA ..................(((...((.(((((.((((.(((((.....))))).((((...)))).....))))))))).))...))).. ( -25.00) >DroSim_CAF1 174355 90 + 1 AACGCCACAAAAACAACAGAAAAAGGCUGCGAAGUGCAAGUACUUUGAGUACUACGAGUAUCUCGGCAUCGCACUCGCAACUGCAUUCGA ..................(((...((.(((((.((((.(((((.....))))).((((...)))).....))))))))).))...))).. ( -25.00) >DroEre_CAF1 183359 90 + 1 AACGCCACAAAAACAACAGAAAAAGGCUGCGAAGUGCAAGUACUUUGAGUACUGCGAGUAUUUCGGCAUCGCACUCGCAACUGCAUUCGA ...(((..................)))..((((.(((((((((.....)))))((((((....((....)).))))))...)))))))). ( -22.87) >DroYak_CAF1 189485 89 + 1 AACGCCACAAAAACAAGAGAA-AAGGCUACGAAGUGCAAGUACUUUGAGUACUACGGGUAUUUCGGCAUCGCACUCGGAACUGCAUUCGA ...(((...............-..)))..((((.(((((((((.....))))).(((((....((....)).)))))....)))))))). ( -19.03) >consensus AACGCCACAAAAACAACAGAAAAAGGCUGCGAAGUGCAAGUACUUUGAGUACUACGAGUAUCUCGGCAUCGCACUCGCAACUGCAUUCGA ..................(((...((.(((((.((((.(((((.....))))).((((...)))).....))))))))).))...))).. (-13.81 = -16.32 + 2.50)

| Location | 15,390,523 – 15,390,620 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 79.93 |
| Mean single sequence MFE | -25.82 |
| Consensus MFE | -12.12 |
| Energy contribution | -13.26 |
| Covariance contribution | 1.14 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.47 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.724380 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15390523 97 + 20766785 AGAAAAAGGCUGCGAAGUGCAAGUACUUUGAGUACUACGAGUAUCUCGGCAUCGCACUCGCAACUGCAUUCGAUAGAUACUCGCAACCACAAACAAG .......((.((((((((((.(((((.....))))).((((...)))).....))))).((....)).............))))).))......... ( -26.30) >DroPse_CAF1 225060 77 + 1 AGCCA-----------------CAACUGCGAG---UGCGAGUAUCUUGUCCUCGCACUCGCAACUGCAUCCGAUAGAUACUCGCAACCGCAAACAAG .....-----------------....((((..---(((((((((((.(((...(((........)))....))))))))))))))..))))...... ( -27.60) >DroSec_CAF1 196100 97 + 1 AGAAAAAGGCUGCGAAGUGCAAGUACUUUGAGUACUACGAGUAUCUCGGCAUCGCACUCGCAACUGAAUUCGAUAGAUACUCGCAACCACAAACAAG .......((.(((((((((....))))))(((((((((((((...((((....((....))..))))))))).))).)))))))).))......... ( -25.60) >DroEre_CAF1 183376 97 + 1 AGAAAAAGGCUGCGAAGUGCAAGUACUUUGAGUACUGCGAGUAUUUCGGCAUCGCACUCGCAACUGCAUUCGAUAGAUACUCGCAACCACAAACAAG ........(((.(((((((....))))))))))..((((((((((((((((..((....))...)))...)))..))))))))))............ ( -26.90) >DroYak_CAF1 189502 96 + 1 AGAA-AAGGCUACGAAGUGCAAGUACUUUGAGUACUACGGGUAUUUCGGCAUCGCACUCGGAACUGCAUUCGAUAGAUACUCGCAACCACAAACAAG ....-...(((.(((((((....))))))))))....((((((((((((.((.(((........)))))))))..)))))))).............. ( -20.90) >DroPer_CAF1 184985 77 + 1 AGCCA-----------------CAACUGCGAG---UGCGAGUAUCUUGUCCUCGCACUCGCAACUGCAUCCGAUAGAUACUCGCAACCGCAAACAAG .....-----------------....((((..---(((((((((((.(((...(((........)))....))))))))))))))..))))...... ( -27.60) >consensus AGAAAAAGGCUGCGAAGUGCAAGUACUUUGAGUACUACGAGUAUCUCGGCAUCGCACUCGCAACUGCAUUCGAUAGAUACUCGCAACCACAAACAAG .....................(((((.....))))).(((((((((.......(((........))).......))))))))).............. (-12.12 = -13.26 + 1.14)

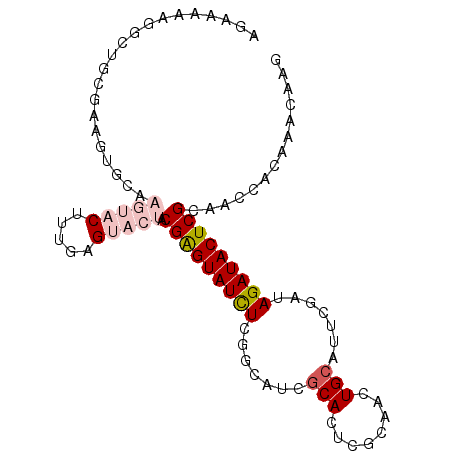

| Location | 15,390,523 – 15,390,620 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 79.93 |
| Mean single sequence MFE | -31.90 |
| Consensus MFE | -17.49 |
| Energy contribution | -18.02 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.98 |
| Structure conservation index | 0.55 |
| SVM decision value | 1.44 |
| SVM RNA-class probability | 0.953794 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15390523 97 - 20766785 CUUGUUUGUGGUUGCGAGUAUCUAUCGAAUGCAGUUGCGAGUGCGAUGCCGAGAUACUCGUAGUACUCAAAGUACUUGCACUUCGCAGCCUUUUUCU .........(((((((((((((..(((...(((.((((....)))))))))))))))))(((((((.....)))).))).....))))))....... ( -32.30) >DroPse_CAF1 225060 77 - 1 CUUGUUUGCGGUUGCGAGUAUCUAUCGGAUGCAGUUGCGAGUGCGAGGACAAGAUACUCGCA---CUCGCAGUUG-----------------UGGCU .........(((..((((((((.....)))))..(((((((((((((.........))))))---))))))))))-----------------..))) ( -35.90) >DroSec_CAF1 196100 97 - 1 CUUGUUUGUGGUUGCGAGUAUCUAUCGAAUUCAGUUGCGAGUGCGAUGCCGAGAUACUCGUAGUACUCAAAGUACUUGCACUUCGCAGCCUUUUUCU .........((((((((((((((((((.((((......)))).))))....))))))))(((((((.....)))).))).....))))))....... ( -32.40) >DroEre_CAF1 183376 97 - 1 CUUGUUUGUGGUUGCGAGUAUCUAUCGAAUGCAGUUGCGAGUGCGAUGCCGAAAUACUCGCAGUACUCAAAGUACUUGCACUUCGCAGCCUUUUUCU .........((((((((((((...(((...(((.((((....)))))))))).))))))(((((((.....)))).))).....))))))....... ( -31.10) >DroYak_CAF1 189502 96 - 1 CUUGUUUGUGGUUGCGAGUAUCUAUCGAAUGCAGUUCCGAGUGCGAUGCCGAAAUACCCGUAGUACUCAAAGUACUUGCACUUCGUAGCCUU-UUCU .........(((((((((((((....).)))).......((((((((((.(......).)))((((.....)))))))))))))))))))..-.... ( -23.80) >DroPer_CAF1 184985 77 - 1 CUUGUUUGCGGUUGCGAGUAUCUAUCGGAUGCAGUUGCGAGUGCGAGGACAAGAUACUCGCA---CUCGCAGUUG-----------------UGGCU .........(((..((((((((.....)))))..(((((((((((((.........))))))---))))))))))-----------------..))) ( -35.90) >consensus CUUGUUUGUGGUUGCGAGUAUCUAUCGAAUGCAGUUGCGAGUGCGAUGCCGAGAUACUCGCAGUACUCAAAGUACUUGCACUUCGCAGCCUUUUUCU ....((((.(..(((((((((((......((((.(....).))))......)))))))))))...).)))).......................... (-17.49 = -18.02 + 0.53)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:27:10 2006