
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
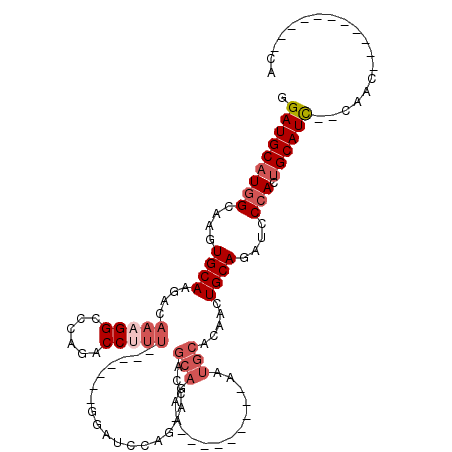
| Location | 15,387,503 – 15,387,593 |
| Length | 90 |
| Max. P | 0.945949 |

| Location | 15,387,503 – 15,387,593 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.33 |
| Mean single sequence MFE | -28.37 |
| Consensus MFE | -13.16 |
| Energy contribution | -14.52 |
| Covariance contribution | 1.36 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.78 |
| Structure conservation index | 0.46 |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.945949 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
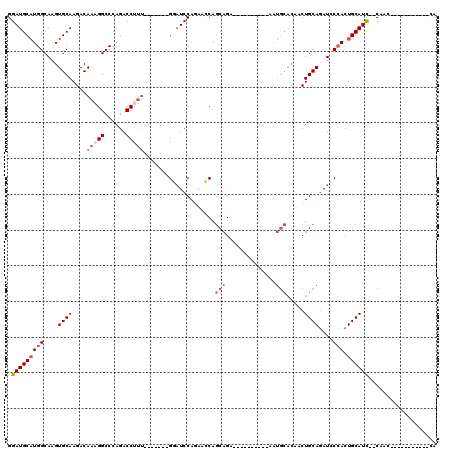
>2R_DroMel_CAF1 15387503 90 + 20766785 GGAUGCAUGGCAAGUGCAAGACAAAGGCCCAGACCUUU-------GGAUCCAGACCCAGCAGA----------AAUGCACAACUGCAGAUCCCACUGCAUU--CAAC-----------CA ((((((((((....((((...((((((......)))))-------)............(((..----------..))).....))))....))).))))))--)...-----------.. ( -28.00) >DroSec_CAF1 193118 90 + 1 GGAUGCAUGGCAAGUGCAAGACAAAGGCCCAGACCUUU-------GGAUCCAGAACCAGCAGA----------AAUGCACAACUGCAGAUCCCACUGCAUC--CAAC-----------CA ((((((((((....((((...((((((......)))))-------)............(((..----------..))).....))))....))).))))))--)...-----------.. ( -30.90) >DroSim_CAF1 171345 90 + 1 GGAUGCAUGGCAAGUGCAAGACAAAGGCCCAGACCUUU-------GGAUCCAGAACCAGCAGA----------AAUGCACAACUGCAGAUCCCACUGCAUC--CAAC-----------CA ((((((((((....((((...((((((......)))))-------)............(((..----------..))).....))))....))).))))))--)...-----------.. ( -30.90) >DroEre_CAF1 180323 90 + 1 GGAUGCAUGGCAAGUGCAAGACAAAGGCCCAGACCCCU-------GCAUCCAGACCCAGCAGA----------GAUGCACAACUGCAGAUCCCACUGCAUC--CGAC-----------CA ((((((((((....((((......(((........)))-------((((((..........).----------))))).....))))....))).))))))--)...-----------.. ( -25.40) >DroYak_CAF1 186328 111 + 1 GGAUGCAUGGCAAGUGCAAGACAAAGGCCCAGACCUUU-------GGAUCCACAACCAGCAGAACCAGCUGAAAAUGCACAACUGCAGAUCCCACUGCAUC--CGACCCAUCCACAUCCA (((((.((((...(((((...((((((......)))))-------)..........((((.......))))....)))))...(((((......)))))..--....))))...))))). ( -34.50) >DroAna_CAF1 168194 92 + 1 GGAUGCCUGGCAAGUGCAAGACAAAGGCCCAGACCCUGCCACAUCGACUCCAUCUUCAACUGG-----------------UACUGCAACUGCAACUGCAUCGUCGCC-----------UG .(((((...(((..((((..((...(((.........))).....((........)).....)-----------------)..))))..)))....)))))......-----------.. ( -20.50) >consensus GGAUGCAUGGCAAGUGCAAGACAAAGGCCCAGACCUUU_______GGAUCCAGAACCAGCAGA__________AAUGCACAACUGCAGAUCCCACUGCAUC__CAAC___________CA .(((((((((....((((....(((((......)))))....................(((..............))).....))))....))).))))))................... (-13.16 = -14.52 + 1.36)

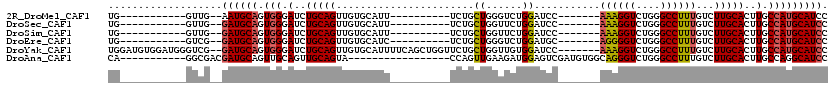
| Location | 15,387,503 – 15,387,593 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.33 |
| Mean single sequence MFE | -33.33 |
| Consensus MFE | -19.25 |
| Energy contribution | -19.42 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.58 |
| SVM decision value | -0.00 |
| SVM RNA-class probability | 0.532356 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
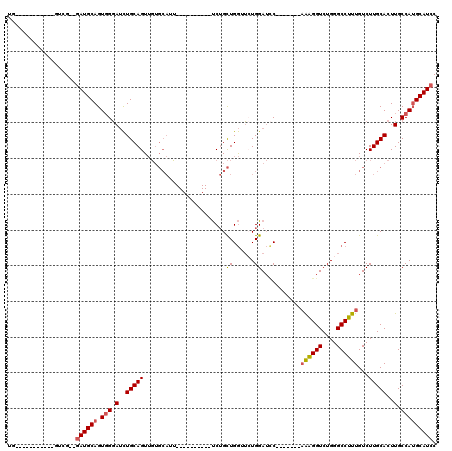
>2R_DroMel_CAF1 15387503 90 - 20766785 UG-----------GUUG--AAUGCAGUGGGAUCUGCAGUUGUGCAUU----------UCUGCUGGGUCUGGAUCC-------AAAGGUCUGGGCCUUUGUCUUGCACUUGCCAUGCAUCC ((-----------((..--..(((((......)))))...(((((..----------.(....(((((..((((.-------...))))..)))))..)...)))))..))))....... ( -31.30) >DroSec_CAF1 193118 90 - 1 UG-----------GUUG--GAUGCAGUGGGAUCUGCAGUUGUGCAUU----------UCUGCUGGUUCUGGAUCC-------AAAGGUCUGGGCCUUUGUCUUGCACUUGCCAUGCAUCC ..-----------...(--(((((((..(((..((((....)))).)----------))..)((((..((.(..(-------((((((....)))))))...).))...))))))))))) ( -31.80) >DroSim_CAF1 171345 90 - 1 UG-----------GUUG--GAUGCAGUGGGAUCUGCAGUUGUGCAUU----------UCUGCUGGUUCUGGAUCC-------AAAGGUCUGGGCCUUUGUCUUGCACUUGCCAUGCAUCC ..-----------...(--(((((((..(((..((((....)))).)----------))..)((((..((.(..(-------((((((....)))))))...).))...))))))))))) ( -31.80) >DroEre_CAF1 180323 90 - 1 UG-----------GUCG--GAUGCAGUGGGAUCUGCAGUUGUGCAUC----------UCUGCUGGGUCUGGAUGC-------AGGGGUCUGGGCCUUUGUCUUGCACUUGCCAUGCAUCC ..-----------...(--((((((.(((.(..(((((....(((..----------..))).(((((..(((..-------....)))..))))).....)))))..).)))))))))) ( -34.30) >DroYak_CAF1 186328 111 - 1 UGGAUGUGGAUGGGUCG--GAUGCAGUGGGAUCUGCAGUUGUGCAUUUUCAGCUGGUUCUGCUGGUUGUGGAUCC-------AAAGGUCUGGGCCUUUGUCUUGCACUUGCCAUGCAUCC .(((((((....(((.(--(.(((((..(((((..(((..(.(((...((....))...))))..)))..)))))-------((((((....))))))...))))))).))).))))))) ( -42.00) >DroAna_CAF1 168194 92 - 1 CA-----------GGCGACGAUGCAGUUGCAGUUGCAGUA-----------------CCAGUUGAAGAUGGAGUCGAUGUGGCAGGGUCUGGGCCUUUGUCUUGCACUUGCCAGGCAUCC ..-----------.(((((..(((....))))))))....-----------------(((........)))....(((((((((((((..((((....)))).)).))))))..))))). ( -28.80) >consensus UG___________GUCG__GAUGCAGUGGGAUCUGCAGUUGUGCAUU__________UCUGCUGGUUCUGGAUCC_______AAAGGUCUGGGCCUUUGUCUUGCACUUGCCAUGCAUCC ...................((((((.(((.(..(((((.......................((......))...........((((((....))))))...)))))..).))))))))). (-19.25 = -19.42 + 0.17)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:27:07 2006