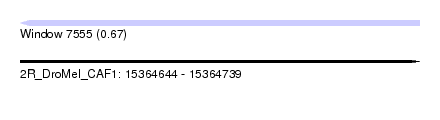
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 15,364,644 – 15,364,739 |
| Length | 95 |
| Max. P | 0.668321 |

| Location | 15,364,644 – 15,364,739 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 80.22 |
| Mean single sequence MFE | -17.12 |
| Consensus MFE | -8.51 |
| Energy contribution | -9.04 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.668321 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
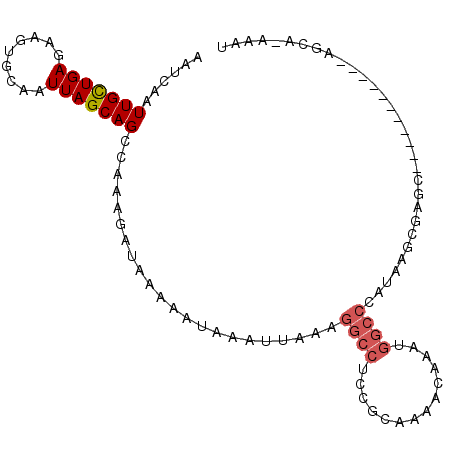
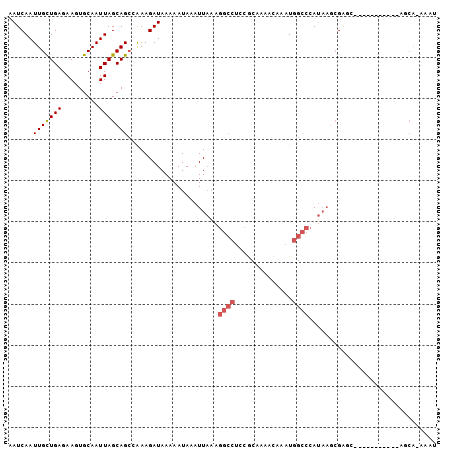
>2R_DroMel_CAF1 15364644 95 - 20766785 AAUCAAUUGCUGAGAACUGCAAUUAGCAGCCAAAGAUAAAAAUAAAUUAAAGGCCUCCACAUAACAAAUGGCCCAUAAUCGGGC--------CACAGCG-AAAU ......((((((....((((.....)))).......................................((((((......))))--------)))))))-)... ( -24.10) >DroPse_CAF1 198420 92 - 1 AAUCAAUUGCUGAGAAGUGCAAUUAGCAGCCAAAGAUAAAAAUAAAUUAAAGGCCUCCGCCAAACAAAUGGCCCAUAAGCGAGC-----------AGCA-AAAU ......((((((.....(((........(((...(((........)))...)))....((((......))))......)))..)-----------))))-)... ( -18.20) >DroGri_CAF1 193552 82 - 1 AAUCAAUUGUUGAGAAGUGCAAUUAGCAGCAAAAGAUAAAAAUAAAUUCACA-----AACAAAACAAAUGGUCCAAC----CAC--------CCAACAA----- .(((..((((((..((......))..))))))..)))...............-----...........(((.....)----)).--------.......----- ( -9.10) >DroWil_CAF1 340852 87 - 1 AAUCAAUUGCUGAGAAGUGCAAUUAGCAGGAAAAGAUAAAAAUAAAUUAAAGGCCUCCGCAAAACAAAUGGCCCAUAAGCG----------------CA-AAAC .(((..(((((((.........))))))).....)))..............((((..............))))........----------------..-.... ( -12.04) >DroAna_CAF1 143555 104 - 1 AAUCAAUUGCUGAGAAGUGCAAUUAGCAGCCGAAGAUAAAAAUAAAUUAAAGGCCUCCGCAUAACAAAUGGCCCAUAAGCGGACAGCAACUGCACAGCACAAAU .......(((((.(.(((((........(((...(((........)))...))).(((((..................)))))..)).))).).)))))..... ( -21.07) >DroPer_CAF1 157323 92 - 1 AAUCAAUUGCUGAGAAGUGCAAUUAGCAGCCAAAGAUAAAAAUAAAUUAAAGGCCUCCGCCAAACAAAUGGCCCAUAAGCGAGC-----------AGCA-AAAU ......((((((.....(((........(((...(((........)))...)))....((((......))))......)))..)-----------))))-)... ( -18.20) >consensus AAUCAAUUGCUGAGAAGUGCAAUUAGCAGCCAAAGAUAAAAAUAAAUUAAAGGCCUCCGCAAAACAAAUGGCCCAUAAGCGAGC___________AGCA_AAAU ......(((((((.........)))))))......................((((..............))))............................... ( -8.51 = -9.04 + 0.53)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:27:00 2006