| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
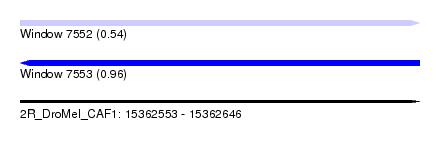
| Location | 15,362,553 – 15,362,646 |
| Length | 93 |
| Max. P | 0.963803 |

| Location | 15,362,553 – 15,362,646 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 79.42 |
| Mean single sequence MFE | -19.56 |
| Consensus MFE | -11.65 |
| Energy contribution | -11.40 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.535763 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
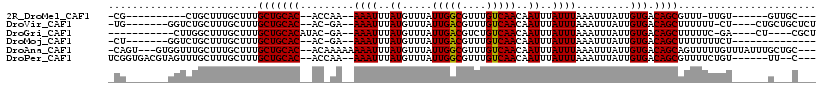
>2R_DroMel_CAF1 15362553 93 + 20766785 -CG----------CUGCUUUGCUUUGCUGCAC--ACCAA--AAAUUUAUGUUUAUUGGCGUUUGUCAACAAUUUAUUUAAAUUUAUUGUGACAGCGUUU-UUGU------GUUGC--- -.(----------(......))...((.((((--(((((--((((....)))).))))((((.((((.((((..((....))..))))))))))))...-.)))------)).))--- ( -19.50) >DroVir_CAF1 217978 100 + 1 -UG-------GGUCUGCUUUGCUUUGCUGCAC--AC-GA--AAAUUUAUGUUUAUUGACGUUUGUCAACAAUUUAUUUAAAUUUAUUGUGACAGCUUUUUU-CU----CUGCUGCUCU -.(-------(((..((........(((((((--(.-..--(((((((...((.(((((....))))).))......)))))))..)))).))))......-..----..)).)))). ( -20.17) >DroGri_CAF1 190470 95 + 1 -----------CUUGGCUUUGCUUUGCUGCACAUAC-GA--AAAUUUAUGUUUAUUGACGUCUGUCAACAAUUUAUUUAAAUUUAUUGUGACAGCUUUUUC-GA----CU----CGCU -----------...(((..(((......)))....(-((--(((...(((((....)))))((((((.((((..((....))..))))))))))..)))))-).----..----.))) ( -19.10) >DroMoj_CAF1 230798 91 + 1 -CU-------GGUCUGCUUUGCUUUGCUGCAC--AC-GA--AAAUUUAUGUUUAUUGACGUUUGUCAACAAUUUAUUUAAAUUUAUUGUGACAGCUUUUUUUCU-------------- -..-------...............(((((((--(.-..--(((((((...((.(((((....))))).))......)))))))..)))).)))).........-------------- ( -15.50) >DroAna_CAF1 141409 109 + 1 -CAGU---GUGGUUUGCUUUGCUUUGCUGCAC--ACAAAAAAAAUUUAUGUUUAUUGGCGUUUGUCAACAAUUUAUUUAAAUUUAUUGUGACAGCAGUUUUUGUUUAUUUGCUGC--- -..((---(..((..((...))...))..)))--((((...(((((((.((..((((...........))))..)).))))))).))))..((((((...........)))))).--- ( -21.00) >DroPer_CAF1 155014 103 + 1 UCGGUGACGUAGUUUGCUUUGCUUUGCUGCAC--ACCAA--AAAUUUAUGUUUAUUGGCGUUUGUCAACAAUUUAUUUAAAUUUAUUGUGACAGCGUUUUCUGU------UU--C--- ..((((..(((((..((...))...))))).)--)))..--...............((((((.((((.((((..((....))..))))))))))))))......------..--.--- ( -22.10) >consensus _CG_______GGUCUGCUUUGCUUUGCUGCAC__AC_AA__AAAUUUAUGUUUAUUGACGUUUGUCAACAAUUUAUUUAAAUUUAUUGUGACAGCUUUUUUUGU______G___C___ .........................(((((((.........((((..((.....(((((....)))))..))..)))).........))).))))....................... (-11.65 = -11.40 + -0.25)


| Location | 15,362,553 – 15,362,646 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 79.42 |
| Mean single sequence MFE | -19.26 |
| Consensus MFE | -10.90 |
| Energy contribution | -10.90 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.88 |
| Structure conservation index | 0.57 |
| SVM decision value | 1.56 |
| SVM RNA-class probability | 0.963803 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
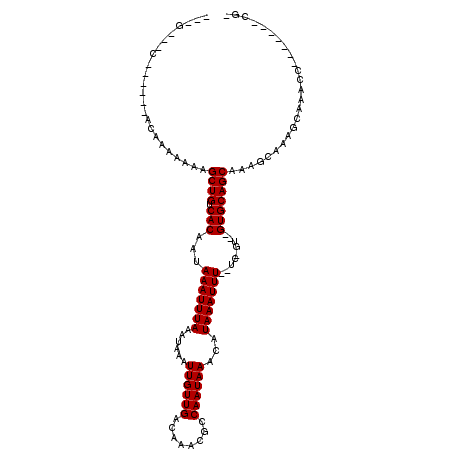
>2R_DroMel_CAF1 15362553 93 - 20766785 ---GCAAC------ACAA-AAACGCUGUCACAAUAAAUUUAAAUAAAUUGUUGACAAACGCCAAUAAACAUAAAUUU--UUGGU--GUGCAGCAAAGCAAAGCAG----------CG- ---.....------....-...((((((.......(((((....)))))((((....(((((((.............--)))))--)).))))........))))----------))- ( -19.32) >DroVir_CAF1 217978 100 - 1 AGAGCAGCAG----AG-AAAAAAGCUGUCACAAUAAAUUUAAAUAAAUUGUUGACAAACGUCAAUAAACAUAAAUUU--UC-GU--GUGCAGCAAAGCAAAGCAGACC-------CA- ..........----..-......((((.((((..(((((((......((((((((....))))))))...)))))))--..-.)--)))))))...............-------..- ( -21.10) >DroGri_CAF1 190470 95 - 1 AGCG----AG----UC-GAAAAAGCUGUCACAAUAAAUUUAAAUAAAUUGUUGACAGACGUCAAUAAACAUAAAUUU--UC-GUAUGUGCAGCAAAGCAAAGCCAAG----------- ....----..----..-......((((.((((..(((((((......((((((((....))))))))...)))))))--..-...))))))))..............----------- ( -18.60) >DroMoj_CAF1 230798 91 - 1 --------------AGAAAAAAAGCUGUCACAAUAAAUUUAAAUAAAUUGUUGACAAACGUCAAUAAACAUAAAUUU--UC-GU--GUGCAGCAAAGCAAAGCAGACC-------AG- --------------.........((((.((((..(((((((......((((((((....))))))))...)))))))--..-.)--)))))))...............-------..- ( -21.10) >DroAna_CAF1 141409 109 - 1 ---GCAGCAAAUAAACAAAAACUGCUGUCACAAUAAAUUUAAAUAAAUUGUUGACAAACGCCAAUAAACAUAAAUUUUUUUUGU--GUGCAGCAAAGCAAAGCAAACCAC---ACUG- ---.(((...............(((((((((((.(((((((......((((((........))))))...)))))))...))))--).))))))..((...)).......---.)))- ( -18.10) >DroPer_CAF1 155014 103 - 1 ---G--AA------ACAGAAAACGCUGUCACAAUAAAUUUAAAUAAAUUGUUGACAAACGCCAAUAAACAUAAAUUU--UUGGU--GUGCAGCAAAGCAAAGCAAACUACGUCACCGA ---(--(.------..((.....(((((..((((((.((.....)).))))))....(((((((.............--)))))--)))))))...((...))...))...))..... ( -17.32) >consensus ___G___C______ACAAAAAAAGCUGUCACAAUAAAUUUAAAUAAAUUGUUGACAAACGCCAAUAAACAUAAAUUU__UC_GU__GUGCAGCAAAGCAAAGCAAACC_______CG_ .......................((((.(((...(((((((......((((((........))))))...))))))).........)))))))......................... (-10.90 = -10.90 + 0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:26:58 2006