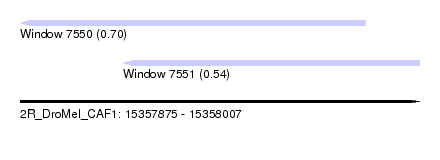
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 15,357,875 – 15,358,007 |
| Length | 132 |
| Max. P | 0.696838 |

| Location | 15,357,875 – 15,357,989 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.44 |
| Mean single sequence MFE | -23.23 |
| Consensus MFE | -19.30 |
| Energy contribution | -19.63 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.696838 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
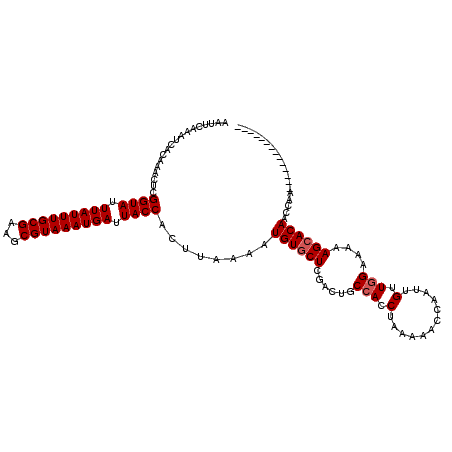
>2R_DroMel_CAF1 15357875 114 - 20766785 AAUUCAAAUCACAAACUCGGUAUUUAUUUGCGAAGCGUAAAUGAUUACCACUUAAAAUGUGCUCGACUGCCACCUAAAAACCAAUUGUUGGAAAAAGCACAACCAACCGAAUGG------ .((((.............((((.(((((((((...))))))))).))))........((((((......(((.(............).)))....)))))).......))))..------ ( -24.30) >DroVir_CAF1 212126 106 - 1 AAUUCAAAUCACAAACUCGGUAUUUAUUUGCGAAGCGUAAAUGAUUACCACUUAAAAUGUGCUCGACUGCCACCUAAAAACCAAUUGUUGGGAAAAGCACAACCGA-------------- ..................((((.(((((((((...))))))))).))))........((((((....(.(((.(............).))).)..)))))).....-------------- ( -21.70) >DroGri_CAF1 183466 106 - 1 AAUUCAAAUCACAAACUCGGUAUUUAUUUGCGAAGCGUAAAUGAUUACCACUUAAAAUGUGCUCGACUGCCACCUAAAAACCAAUUGUUGGGAAAAGGACAACCGA-------------- ..................((((.(((((((((...))))))))).))))........((.((......))))(((.....(((.....)))....)))........-------------- ( -19.50) >DroWil_CAF1 322623 119 - 1 AAUUCAAAUCACAAACUCGGUAUUUAUUUGCGAAGCGUAAAUGAUUACCACUUAAAAUGUGCUCGACUGCCACCUAAAAACCAAUUGUUGGAAAAAGCACAACGAACGGAGUGAACAGA- ........((((......((((.(((((((((...))))))))).))))........((((((......(((.(............).)))....)))))).........)))).....- ( -26.60) >DroMoj_CAF1 222555 106 - 1 AAUUCAAAUCAGAAACUCGGUAUUUAUUUGCGAAGCGUAAAUGAUUACCACUUAAAAUGUGCUCGACUGCCACCUAAAAACCAAUUGUUAGGAAAAGCACAACCGA-------------- ..................((((.(((((((((...))))))))).))))........((((((.(.....).(((((..........)))))...)))))).....-------------- ( -21.70) >DroAna_CAF1 137161 120 - 1 AAUUCAAAUCACAAACUCGGUAUUUAUUUGCGAAGCGUAAAUGAUUACCACUUAAAAUGUGCUCGACUGCCACCUAAAAACCAAUUGUUGGAAAAAGCACAACCAAGCGAGAGGCCAGAG ...............(((((((.(((((((((...))))))))).))))........((((((......(((.(............).)))....)))))).........)))....... ( -25.60) >consensus AAUUCAAAUCACAAACUCGGUAUUUAUUUGCGAAGCGUAAAUGAUUACCACUUAAAAUGUGCUCGACUGCCACCUAAAAACCAAUUGUUGGAAAAAGCACAACCAA______________ ..................((((.(((((((((...))))))))).))))........((((((......(((.(............).)))....))))))................... (-19.30 = -19.63 + 0.33)

| Location | 15,357,909 – 15,358,007 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.58 |
| Mean single sequence MFE | -22.32 |
| Consensus MFE | -14.90 |
| Energy contribution | -14.90 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.544721 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15357909 98 - 20766785 AAA-----------UGGAAAUG-----------GCAGGAAAAUUCAAAUCACAAACUCGGUAUUUAUUUGCGAAGCGUAAAUGAUUACCACUUAAAAUGUGCUCGACUGCCACCUAAAAA ...-----------.((...((-----------((((((....)).............((((.(((((((((...))))))))).)))).................))))))))...... ( -22.80) >DroVir_CAF1 212152 98 - 1 GUC-----------UGCGAAGGGGGC-----------GAAAAUUCAAAUCACAAACUCGGUAUUUAUUUGCGAAGCGUAAAUGAUUACCACUUAAAAUGUGCUCGACUGCCACCUAAAAA ...-----------.....(((.(((-----------((....)).............((((.(((((((((...))))))))).))))...................))).)))..... ( -21.40) >DroGri_CAF1 183492 104 - 1 GUA-----------UUCAAGCGG-----CAGCAGGGAGAAAAUUCAAAUCACAAACUCGGUAUUUAUUUGCGAAGCGUAAAUGAUUACCACUUAAAAUGUGCUCGACUGCCACCUAAAAA ...-----------.......((-----(((..(((.((....)).............((((.(((((((((...))))))))).))))............)))..)))))......... ( -25.60) >DroWil_CAF1 322662 94 - 1 --------------GGGCAAU------------GAAAGAAAAUUCAAAUCACAAACUCGGUAUUUAUUUGCGAAGCGUAAAUGAUUACCACUUAAAAUGUGCUCGACUGCCACCUAAAAA --------------(((((.(------------(((......))))............((((.(((((((((...))))))))).))))..........)))))................ ( -20.60) >DroMoj_CAF1 222581 109 - 1 GAU-----------GACGAGGGGGGUCCCUGCUGCCAGAAAAUUCAAAUCAGAAACUCGGUAUUUAUUUGCGAAGCGUAAAUGAUUACCACUUAAAAUGUGCUCGACUGCCACCUAAAAA ...-----------...(..((.((((...((..........(((......)))....((((.(((((((((...))))))))).))))...........))..)))).))..)...... ( -24.90) >DroAna_CAF1 137201 109 - 1 AAAUGAAAUGGAGAUGGAAAUG-----------GAAGGAAAAUUCAAAUCACAAACUCGGUAUUUAUUUGCGAAGCGUAAAUGAUUACCACUUAAAAUGUGCUCGACUGCCACCUAAAAA ..........(((.((((....-----------(((......)))...)).))..)))((((.(((((((((...))))))))).))))........((.((......))))........ ( -18.60) >consensus GAA___________UGCAAAUG___________GAAAGAAAAUUCAAAUCACAAACUCGGUAUUUAUUUGCGAAGCGUAAAUGAUUACCACUUAAAAUGUGCUCGACUGCCACCUAAAAA ..........................................................((((.(((((((((...))))))))).))))........((.((......))))........ (-14.90 = -14.90 + 0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:26:56 2006