| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 15,344,583 – 15,344,703 |
| Length | 120 |
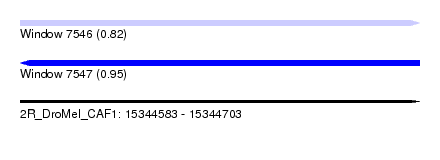
| Max. P | 0.949225 |

| Location | 15,344,583 – 15,344,703 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.56 |
| Mean single sequence MFE | -45.85 |
| Consensus MFE | -22.40 |
| Energy contribution | -23.10 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.42 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.49 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.823296 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
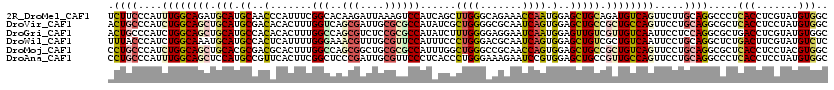
>2R_DroMel_CAF1 15344583 120 + 20766785 UCUUCCCAUUUGGCAGAUGCAUGCAACCCAUUUCGGCACAAGAUUAAAGUCCAUCAGCUUGGGCAGAAACCAAUGGAGCUGCAGAUGUCAGUUCUUGCAGGCCCUCACCUCGUAUGUGGC .....((((((((((..((((.((...(((((((.......)).....(((((......))))).......))))).))))))..))))))....((((((......))).))).)))). ( -30.00) >DroVir_CAF1 190630 120 + 1 ACUGCCCAUCUGGCAGCUGCAUGCGACACACUUUGGUCAGCGAUUGCGCGCCAUAUCGCUGGGGCGCAAUCAGUGGAGCUGCCGCUGCCAGUUCCUGCAGGCGCUCACCUCCUAUGUGGC .((((....((((((((.(((.((..(((((....))....((((((((.(((......))).)))))))).)))..))))).)))))))).....))))..((.(((.......))))) ( -56.20) >DroGri_CAF1 162349 120 + 1 ACUGCCCAUCUGGCAGCUGCAUGCCACACACUUUGGCCAGCGUCUCCGCGCCAUAUCUUUGGGGAGGAAUCAAUGGAGUUGUCGUUGUCAAUUCCUCCAGGCGCUGACCUCGUAUGUGGC .(((((.....)))))......((((((.((...((.((((((((.....(((......)))((((((((((((((.....))))))...)))))))))))))))).))..)).)))))) ( -51.40) >DroWil_CAF1 293780 120 + 1 UUUACCCAUCUGGCAAAUGCAUGCCACUCAUUUUGGGAAACGUUUGCGUUCCAUUUCCCUGGGACGCAAUCAGUGGAGCUGUCGCUGUCAAUUCCUGCAGGCUCUGACUUCGUAUGUCUC ....((((..(((((......))))).......))))..((..((((((((((......))))))))))...))((((((...((...........)).))))))(((.......))).. ( -37.00) >DroMoj_CAF1 197469 120 + 1 CCUGCCCAUCUGGCAGCUGCACGCGACGCACUUUGGCCAGCGGCUGCGCGCCAUUUGGCUGGGCCGCAACCAGUGGAGCUGCCGCUGUCAGUUCCUGCAGGCGCUCACCUCCUACGUGGC (((((....((((((((.(((.((..(((....(((...((((((.((.(((....)))))))))))..))))))..))))).)))))))).....))))).((.(((.......))))) ( -53.70) >DroAna_CAF1 123126 120 + 1 CCUGCCCAUUUGGCAGCUCCAUGCCGUUCACUUCGGCUCCCGAUUGCGUUCCCUCACCCUGGGAAAGAAUCCGUGGAGCUGCCGUUGCCAGUUCCUGCAGGCCCUCACCUCCUAUGUGGC ...((((((..(((((((((((((((.......))))....((((.(.(((((.......))))).))))).)))))))))))...(((.((....)).)))...........))).))) ( -46.80) >consensus ACUGCCCAUCUGGCAGCUGCAUGCCACACACUUUGGCCAGCGAUUGCGCGCCAUAUCCCUGGGAAGCAAUCAGUGGAGCUGCCGCUGUCAGUUCCUGCAGGCGCUCACCUCCUAUGUGGC .((((....((((((((.(((.((..(.......(((..(((....))))))......((((.......)))).)..))))).)))))))).....)))).....(((.......))).. (-22.40 = -23.10 + 0.70)

| Location | 15,344,583 – 15,344,703 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.56 |
| Mean single sequence MFE | -49.43 |
| Consensus MFE | -24.71 |
| Energy contribution | -24.83 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.45 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.50 |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.949225 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
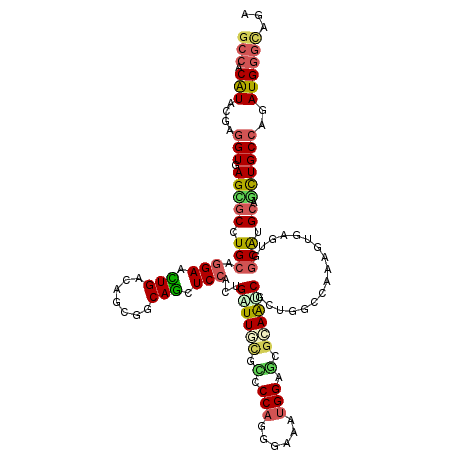
>2R_DroMel_CAF1 15344583 120 - 20766785 GCCACAUACGAGGUGAGGGCCUGCAAGAACUGACAUCUGCAGCUCCAUUGGUUUCUGCCCAAGCUGAUGGACUUUAAUCUUGUGCCGAAAUGGGUUGCAUGCAUCUGCCAAAUGGGAAGA ((.((.((((((((((((((.((((.((.......))))))))(((((..((((......))))..))))))))).))))))))((.....)))).)).....(((.((.....)).))) ( -32.30) >DroVir_CAF1 190630 120 - 1 GCCACAUAGGAGGUGAGCGCCUGCAGGAACUGGCAGCGGCAGCUCCACUGAUUGCGCCCCAGCGAUAUGGCGCGCAAUCGCUGACCAAAGUGUGUCGCAUGCAGCUGCCAGAUGGGCAGU (((.(((.(.((((....)))).).....((((((((.(((((..(((.((((((((.(((......))).))))))))(((......))))))..)).))).))))))))))))))... ( -60.20) >DroGri_CAF1 162349 120 - 1 GCCACAUACGAGGUCAGCGCCUGGAGGAAUUGACAACGACAACUCCAUUGAUUCCUCCCCAAAGAUAUGGCGCGGAGACGCUGGCCAAAGUGUGUGGCAUGCAGCUGCCAGAUGGGCAGU (((((((((..((((((((((.((((((((..(....((....))..)..))))))))..........)))..(....))))))))...))))))))).....((((((.....)))))) ( -56.80) >DroWil_CAF1 293780 120 - 1 GAGACAUACGAAGUCAGAGCCUGCAGGAAUUGACAGCGACAGCUCCACUGAUUGCGUCCCAGGGAAAUGGAACGCAAACGUUUCCCAAAAUGAGUGGCAUGCAUUUGCCAGAUGGGUAAA ..(((.......(((((..((....))..))))).(((((((.....))).)))))))(((((((((((.........))))))))........(((((......)))))..)))..... ( -38.60) >DroMoj_CAF1 197469 120 - 1 GCCACGUAGGAGGUGAGCGCCUGCAGGAACUGACAGCGGCAGCUCCACUGGUUGCGGCCCAGCCAAAUGGCGCGCAGCCGCUGGCCAAAGUGCGUCGCGUGCAGCUGCCAGAUGGGCAGG (((.(((.((((.((..(((.(((((...))).))))).)).)))).(((((.(((((...))).....((((((.((((((......)))).)).)))))).)).)))))))))))... ( -54.90) >DroAna_CAF1 123126 120 - 1 GCCACAUAGGAGGUGAGGGCCUGCAGGAACUGGCAACGGCAGCUCCACGGAUUCUUUCCCAGGGUGAGGGAACGCAAUCGGGAGCCGAAGUGAACGGCAUGGAGCUGCCAAAUGGGCAGG (((.(((.(...((.((..((....))..)).))..)(((((((((((.(((((.(((((.......))))).).)))).)..((((.......)))).))))))))))..))))))... ( -53.80) >consensus GCCACAUACGAGGUGAGCGCCUGCAGGAACUGACAGCGGCAGCUCCACUGAUUGCGCCCCAGGGAAAUGGAGCGCAAUCGCUGGCCAAAGUGAGUGGCAUGCAGCUGCCAGAUGGGCAGA (((.(((....(((.(((((.(((.(((.(((.......))).)))...((((((.(.(((......))).).)))))).................))).)).))))))..))))))... (-24.71 = -24.83 + 0.12)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:26:52 2006