
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 15,328,780 – 15,328,908 |
| Length | 128 |
| Max. P | 0.936890 |

| Location | 15,328,780 – 15,328,873 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 90.61 |
| Mean single sequence MFE | -27.20 |
| Consensus MFE | -22.12 |
| Energy contribution | -23.65 |
| Covariance contribution | 1.53 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.726928 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
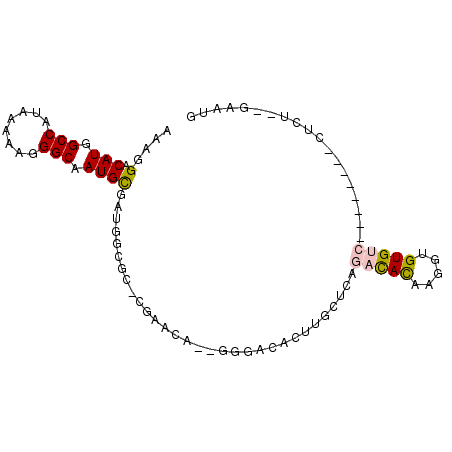
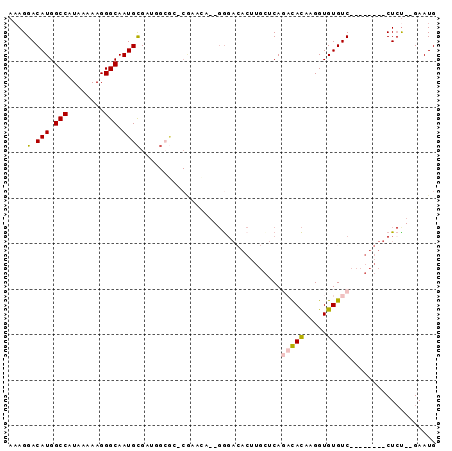
>2R_DroMel_CAF1 15328780 93 - 20766785 GCCCGAACAGGGACACUUGCUCAGACACAAGGUGUGUCUCUGUGUUCUCUGAAUGAAUUGCUGAGUAACCAGUAAUGAAGUAAAACUGUUCAA ....((((((((((((......((((((.....))))))..)))))).........(((((((......))))))).........)))))).. ( -28.60) >DroSec_CAF1 134772 93 - 1 GCCCGAGCAGGGACACUUGCUCAGACACAAGGUGUGUCUCUGUGUUCUCUGAAUGAAUUGCUGAGUAACCAGUAAUGAAGUAAAACUGUUCAA ....((((((((((((......((((((.....))))))..)))))).........(((((((......))))))).........)))))).. ( -28.60) >DroSim_CAF1 112516 93 - 1 GCCCGAGCAGGGACACUUGCUCAGACACAAGGUGUGUCUCUGUGUUCUCUGAAUGAAUUGCUGAGUAACCAGUAAUGAAGUAAAACUGUUCAA ....((((((((((((......((((((.....))))))..)))))).........(((((((......))))))).........)))))).. ( -28.60) >DroEre_CAF1 121368 93 - 1 GCCCGAGCAGGGACACUUGCUCAGACACAAGGUGUGUCUCUGUGUUCUCCGAAUGAAUUGCUGAGUAACCAGUAAUGAAGUAAAACUGUUCAA ....((((((((((((......((((((.....))))))..)))))).........(((((((......))))))).........)))))).. ( -28.60) >DroYak_CAF1 125682 93 - 1 GCCCGAACAGGGACACUUGCUCAGACACAAGGUGUGUCUCUGUGUUCUCUGAAUGGAUUGCUGAGUAACCAGUAAUGAAGUAAAACUGUUCAA ....((((((((((((......((((((.....))))))..)))))).........(((((((......))))))).........)))))).. ( -28.70) >DroAna_CAF1 108490 78 - 1 -------GAGGGACACAUCUACAGAUACAAGGUGUGUC--------CUCUGAAUGAAUUGCUGAGUAACCAAUAAUGAAGUAAAACUGUUCGA -------.((((((((((((.........)))))))))--------).))(((((..(((((..((........))..)))))...))))).. ( -20.10) >consensus GCCCGAGCAGGGACACUUGCUCAGACACAAGGUGUGUCUCUGUGUUCUCUGAAUGAAUUGCUGAGUAACCAGUAAUGAAGUAAAACUGUUCAA ....((((((((((((......((((((.....))))))..)))))).........(((((((......))))))).........)))))).. (-22.12 = -23.65 + 1.53)



| Location | 15,328,818 – 15,328,908 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 72.12 |
| Mean single sequence MFE | -26.18 |
| Consensus MFE | -8.53 |
| Energy contribution | -8.67 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.33 |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.936890 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15328818 90 - 20766785 AAAGGACAUGGCCAUAAAAAGGGCAAUGCGAUGGCGCCCGAACA--GGGACACUUGCUCAGACACAAGGUGUGUCUCUGUGUUCUCU--GAAUG ..(((((((((.........((((((.((....))((((.....--))).)..))))))((((((.....)))))))))))))))..--..... ( -28.70) >DroVir_CAF1 169040 76 - 1 AAAGGACAUGGCCAUAAAAAGGGCAAUGCGAGCGAGG-CAAGUGCUGGC---------CAACUACAAGGUGUGUC--------CUCUGUGAAUG ..((((((((((((........(((.(((.......)-))..)))))))---------).(((....))))))))--------))......... ( -21.10) >DroGri_CAF1 141430 72 - 1 AAAGGACAUGGCCAUAAAAAGGGCAAUGCGAGAGCAG-CAGCAGC--AG---------CAACUAUAAGGUGUGUC--------CUGU--GAAUG ..((((((((.((.........((..(((....))))-).((...--.)---------)........))))))))--------))..--..... ( -20.10) >DroEre_CAF1 121406 90 - 1 AAAGGACAUGGCCAUAAAAAGGGCAAUGCGAUGGCGCCCGAGCA--GGGACACUUGCUCAGACACAAGGUGUGUCUCUGUGUUCUCC--GAAUG ..(((((((((.........((((...((....))))))(((((--((....)))))))((((((.....)))))))))))))))..--..... ( -34.50) >DroYak_CAF1 125720 90 - 1 AAAGGACAUGGCCAUAAAAAGGGCAAUGCGAUGGCGCCCGAACA--GGGACACUUGCUCAGACACAAGGUGUGUCUCUGUGUUCUCU--GAAUG ..(((((((((.........((((((.((....))((((.....--))).)..))))))((((((.....)))))))))))))))..--..... ( -28.70) >DroAna_CAF1 108528 69 - 1 A-AGGACAUGGCCAUAAAAAGGGCAAUGUG------------GA--GGGACACAUCUACAGAUACAAGGUGUGUC--------CUCU--GAAUG .-...((((.(((........))).))))(------------((--((.((((((((.........)))))))))--------))))--..... ( -24.00) >consensus AAAGGACAUGGCCAUAAAAAGGGCAAUGCGAUGGCGC_CGAACA__GGGACACUUGCUCAGACACAAGGUGUGUC________CUCU__GAAUG ....(.(((.(((........))).))))...............................(((((.....)))))................... ( -8.53 = -8.67 + 0.14)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:26:49 2006