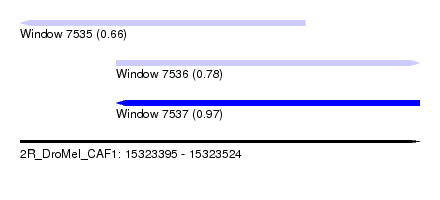
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 15,323,395 – 15,323,524 |
| Length | 129 |
| Max. P | 0.973059 |

| Location | 15,323,395 – 15,323,487 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 79.07 |
| Mean single sequence MFE | -26.01 |
| Consensus MFE | -13.99 |
| Energy contribution | -13.88 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.664712 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
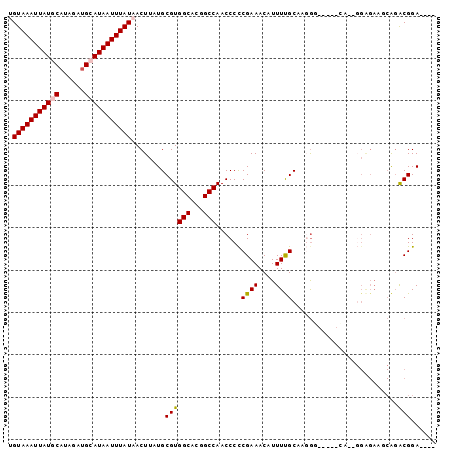
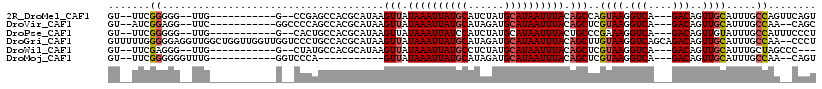
>2R_DroMel_CAF1 15323395 92 - 20766785 UGUAAAUUAUGCAUAGAUGCAUAAUUUAUAACUUAUGCGUGGCUCGGCCAACCCCCGAAACAUUUUGCCGGGG-----CAAAGGAGAUGCAGACGGA---- (((((((((((((....))))))))))))).......(((.((((..((...(((((...........)))))-----....)).)).))..)))..---- ( -28.50) >DroPse_CAF1 154931 97 - 1 AGUAAAUUAUGCAUAGAUGGAUAAUUUAUAACUUAUGCGUGGCAGUGCCAACCCCCGAAACAUUUUGCAUGGUACGCUGACGGGGGAAUCAGGCGGA---- .(((((((((.((....)).)))))))))......(((.((((...)))).((((((...((....((.......)))).))))))......)))..---- ( -24.80) >DroEre_CAF1 116050 91 - 1 UGUAAAUUAUGCAUAGAUGCAUAAUUUAUAACUUAUGCGUGGCUC-GCCAACCCCCGAAACAUUUUGCCCUGG-----CAAAGGAGAUGCAGACGGA---- (((((((((((((....))))))))))))).((..(((((((...-.)).............((((((....)-----)))))...)))))...)).---- ( -25.50) >DroWil_CAF1 257462 98 - 1 UGUAAAUUAUGCAUAGAGGCAUAAUUUAUAACUUAUGCGUGGCAUAGCCAACCCUCGAAACAUUUCGCAAUGCACACGCA---GAGAGACGGACGGACAUU ((((((((((((......)))))))))))).(((.((((((((((.((..................)).)))).))))))---..)))............. ( -28.57) >DroAna_CAF1 103151 80 - 1 UGUAAAUUAUGCAUAGAUGCAUAAUUUAUAACUUAUGCGUGGCUCGGCCAACCCCCGAAACAUUUCGCCAGAG-----------------ACACGGA---- (((((((((((((....))))))))))))).......((((.((((((..................))).)))-----------------.))))..---- ( -23.87) >DroPer_CAF1 113893 97 - 1 AGUAAAUUAUGCAUAGAUGGAUAAUUUAUAACUUAUGCGUGGCAGUGCCAACCCCCGAAACAUUUUGCAUGGUACGCUGACGGGGGAAUCAGGCGGA---- .(((((((((.((....)).)))))))))......(((.((((...)))).((((((...((....((.......)))).))))))......)))..---- ( -24.80) >consensus UGUAAAUUAUGCAUAGAUGCAUAAUUUAUAACUUAUGCGUGGCACGGCCAACCCCCGAAACAUUUUGCAAGGG_____CA__GGAGAAGCAGACGGA____ .(((((((((((......)))))))))))........((((((...)))......((((....)))).........................)))...... (-13.99 = -13.88 + -0.11)

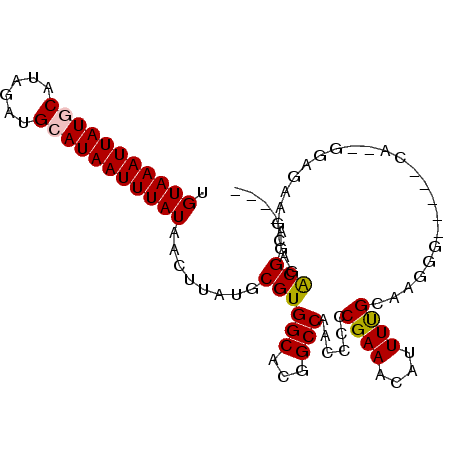
| Location | 15,323,426 – 15,323,524 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 77.43 |
| Mean single sequence MFE | -29.07 |
| Consensus MFE | -15.34 |
| Energy contribution | -15.73 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.777255 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15323426 98 + 20766785 GU--UUCGGGGG--UUG-----------G--CCGAGCCACGCAUAAGUUAUAAAUUAUGCAUCUAUGCAUAAUUUACAGCCAGUAAGGUCA---GACAGUUGCAUUUGCCAGUUCAGU ..--.....(..--(((-----------(--(.((((.((..........(((((((((((....)))))))))))..(((.....)))..---....)).)).)).)))))..)... ( -26.20) >DroVir_CAF1 161056 98 + 1 GU--AUCGGAGG--UUC-----------GGCCCCAGCCACGCAUAAGUUAUAAAUUAUGCAUAGAUGCAUAAUUUACAGCUCGUAAGGUCA---GACAGUUGCAUUUGCCAA--CAGC ..--.......(--(..-----------((((...((...))(..((((.(((((((((((....))))))))))).))))..)..)))).---.)).(((((....).)))--)... ( -24.40) >DroPse_CAF1 154967 98 + 1 GU--UUCGGGGG--UUG-----------G--CACUGCCACGCAUAAGUUAUAAAUUAUCCAUCUAUGCAUAAUUUACUGCCCGAAAGGUCA---GACAGUUGUAUUUGCCAUUUCCCU ..--...(((((--.((-----------(--((.(((.((..........((((((((.((....)).))))))))(((.((....)).))---)...)).)))..))))).))))). ( -29.30) >DroGri_CAF1 134175 116 + 1 GUUUUUGGGGGAGGUUGGCUGGUUGGUUGGUCCCUGCCACGCAUAAGUUAUAAAUUAUGCAUAGAUGCAUAAUUUACAGCUUGUAAGGUCAGCAGACAGUUGCAUUUGCCAA--CCCU .........((.(((((((((((.((......)).)))).(((((((((.(((((((((((....))))))))))).)))))))...(((....)))....))....)))))--)))) ( -40.90) >DroWil_CAF1 257499 95 + 1 GU--UUCGAGGG--UUG-----------G--CUAUGCCACGCAUAAGUUAUAAAUUAUGCCUCUAUGCAUAAUUUACAGCUCGUAAGGUCA---GACAGUUGCAUUUGCUAGCCC--- ..--.....(((--(((-----------(--(.((((.((.....((((.((((((((((......)))))))))).))))(....)....---....)).))))..))))))))--- ( -31.20) >DroMoj_CAF1 164833 89 + 1 GU--UUCGGGGGGUUUG-----------GGUCCCA-----------GUUAUAAAUUAUGCAUAGAUGCAUAAUUUACAGCUCGUAAGGUCA---GACAGUUGCAUUUGCCAA--CAGU ((--((.((((((....-----------..))))(-----------(((.(((((((((((....))))))))))).)))).......)))---))).(((((....).)))--)... ( -22.40) >consensus GU__UUCGGGGG__UUG___________G__CCCAGCCACGCAUAAGUUAUAAAUUAUGCAUAGAUGCAUAAUUUACAGCUCGUAAGGUCA___GACAGUUGCAUUUGCCAA__CAGU .......((.....................................(((.((((((((((......)))))))))).)))..((((.(((....)))..)))).....))........ (-15.34 = -15.73 + 0.39)
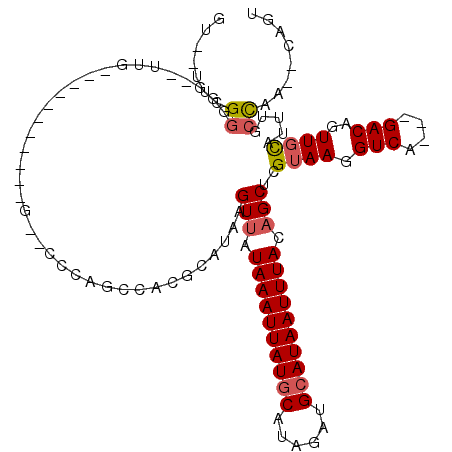
| Location | 15,323,426 – 15,323,524 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 77.43 |
| Mean single sequence MFE | -28.45 |
| Consensus MFE | -13.32 |
| Energy contribution | -15.23 |
| Covariance contribution | 1.92 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.80 |
| Structure conservation index | 0.47 |
| SVM decision value | 1.70 |
| SVM RNA-class probability | 0.973059 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15323426 98 - 20766785 ACUGAACUGGCAAAUGCAACUGUC---UGACCUUACUGGCUGUAAAUUAUGCAUAGAUGCAUAAUUUAUAACUUAUGCGUGGCUCGG--C-----------CAA--CCCCCGAA--AC .......((((..(((((...(((---..........)))(((((((((((((....))))))))))))).....)))))......)--)-----------)).--........--.. ( -24.00) >DroVir_CAF1 161056 98 - 1 GCUG--UUGGCAAAUGCAACUGUC---UGACCUUACGAGCUGUAAAUUAUGCAUCUAUGCAUAAUUUAUAACUUAUGCGUGGCUGGGGCC-----------GAA--CCUCCGAU--AC ..((--((((...(((((...((.---.......))(((.(((((((((((((....))))))))))))).))).)))))(((....)))-----------...--...)))))--). ( -27.10) >DroPse_CAF1 154967 98 - 1 AGGGAAAUGGCAAAUACAACUGUC---UGACCUUUCGGGCAGUAAAUUAUGCAUAGAUGGAUAAUUUAUAACUUAUGCGUGGCAGUG--C-----------CAA--CCCCCGAA--AC .(((...(((((..(((.((((((---(((....)))))))))(((((((.((....)).)))))))...........)))....))--)-----------)).--..)))...--.. ( -30.90) >DroGri_CAF1 134175 116 - 1 AGGG--UUGGCAAAUGCAACUGUCUGCUGACCUUACAAGCUGUAAAUUAUGCAUCUAUGCAUAAUUUAUAACUUAUGCGUGGCAGGGACCAACCAACCAGCCAACCUCCCCCAAAAAC .(((--(((((..(((((...(((....))).....(((.(((((((((((((....))))))))))))).))).)))))((..((......))..)).))))))))........... ( -39.90) >DroWil_CAF1 257499 95 - 1 ---GGGCUAGCAAAUGCAACUGUC---UGACCUUACGAGCUGUAAAUUAUGCAUAGAGGCAUAAUUUAUAACUUAUGCGUGGCAUAG--C-----------CAA--CCCUCGAA--AC ---.(((((((..(((((...((.---.......))(((.((((((((((((......)))))))))))).))).))))).)).)))--)-----------)..--........--.. ( -27.80) >DroMoj_CAF1 164833 89 - 1 ACUG--UUGGCAAAUGCAACUGUC---UGACCUUACGAGCUGUAAAUUAUGCAUCUAUGCAUAAUUUAUAAC-----------UGGGACC-----------CAAACCCCCCGAA--AC ...(--(..(((........)).)---..))....((((.(((((((((((((....))))))))))))).)-----------)(((...-----------.....))).))..--.. ( -21.00) >consensus ACUG__UUGGCAAAUGCAACUGUC___UGACCUUACGAGCUGUAAAUUAUGCAUAGAUGCAUAAUUUAUAACUUAUGCGUGGCAGGG__C___________CAA__CCCCCGAA__AC .........((..(((((...(((....))).....(((.((((((((((((......)))))))))))).))).))))).))................................... (-13.32 = -15.23 + 1.92)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:26:42 2006