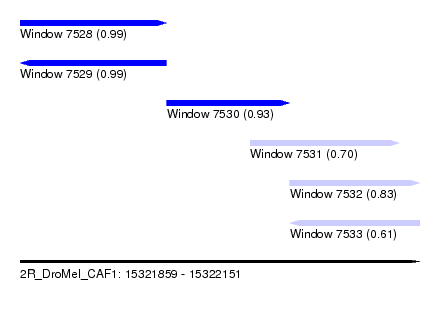
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 15,321,859 – 15,322,151 |
| Length | 292 |
| Max. P | 0.991199 |

| Location | 15,321,859 – 15,321,966 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 78.42 |
| Mean single sequence MFE | -35.44 |
| Consensus MFE | -21.09 |
| Energy contribution | -22.20 |
| Covariance contribution | 1.11 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.23 |
| Structure conservation index | 0.59 |
| SVM decision value | 2.16 |
| SVM RNA-class probability | 0.989291 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
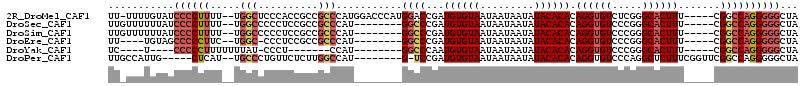
>2R_DroMel_CAF1 15321859 107 + 20766785 UU-UUUUGUAUCCCCUUUU--UGGCUCCCACCGCCGCCCAUGGACCCAUGGACCGAUGUGUAAUAAUAAUAUACACACAGGUGUCUCGGGCACUUU-----CGGCCAGGGGGCUA ..-........(((((...--(((((......(((..(((((....)))))(((..((((((.........))))))..)))......))).....-----.))))))))))... ( -36.00) >DroSec_CAF1 127853 100 + 1 UUGUUUUUUAUCCCCUUUU--UGGCCCCCUCCGCCGCCCAU--------GGCCCGAUGUGUAAUAAUAAUAUACACACAGGUGUCCCGGGCACUUU-----CGGCCAGGGGGCUA ...................--(((((((((..(((((((..--------(((((..((((((.........))))))..)).)))..)))).....-----.))).))))))))) ( -38.60) >DroSim_CAF1 105597 100 + 1 UUGUUUUUUAUCCCCUUUU--UGGCCCCCUCCGCCGCCCAU--------GGCCCGAUGUGUAAUAAUAAUAUACACACAGGUGUCCCGGGCACUUU-----CGGCCAGGGGGCUA ...................--(((((((((..(((((((..--------(((((..((((((.........))))))..)).)))..)))).....-----.))).))))))))) ( -38.60) >DroEre_CAF1 114567 95 + 1 UU----UGUAGCCCCCUUC--UGGC-CCCUCCGCCGCCCAU--------GGCCCGAUGUGUAAUAAUAAUAUACACACAGGUGUCCCGGGCACUUU-----CGGCCAGGGGGCUA ..----..(((((((((..--.(((-(........((((..--------(((((..((((((.........))))))..)).)))..)))).....-----.))))))))))))) ( -42.34) >DroYak_CAF1 118411 86 + 1 UC----U----CCCCCUUUUUUUAU-CCCU-------CCAU--------GGCCCAAUGUGUAAUAAUAAUAUACACACAGGUGUCCCGGGCACUUU-----CGGCCAGGGGGCUA ..----.----..............-.(((-------((.(--------((((...((((((.........)))))).((((((.....)))))).-----.))))))))))... ( -27.40) >DroPer_CAF1 112105 99 + 1 UUGCCAUUG-----CUCAU--UGCCCUGUUCUCUUGGCCAU--------G-UCCGAUGUGUAAUAAUAAUAUACACACAGGUGUCCCAGGCUCUUUCGGUUCGGCCAGGGGGCUA ..(((...(-----(....--.)).......((((((((..--------.-.((((((((((.........)))))).(((.((.....)).)))))))...))))))))))).. ( -29.70) >consensus UU_UUUUUUAUCCCCUUUU__UGGCCCCCUCCGCCGCCCAU________GGCCCGAUGUGUAAUAAUAAUAUACACACAGGUGUCCCGGGCACUUU_____CGGCCAGGGGGCUA ...........((((((.....(((..........)))...........((((...((((((.........)))))).((((((.....)))))).......))))))))))... (-21.09 = -22.20 + 1.11)
| Location | 15,321,859 – 15,321,966 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 78.42 |
| Mean single sequence MFE | -38.55 |
| Consensus MFE | -20.69 |
| Energy contribution | -22.22 |
| Covariance contribution | 1.53 |
| Combinations/Pair | 1.15 |
| Mean z-score | -3.39 |
| Structure conservation index | 0.54 |
| SVM decision value | 2.25 |
| SVM RNA-class probability | 0.991199 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15321859 107 - 20766785 UAGCCCCCUGGCCG-----AAAGUGCCCGAGACACCUGUGUGUAUAUUAUUAUUACACAUCGGUCCAUGGGUCCAUGGGCGGCGGUGGGAGCCA--AAAAGGGGAUACAAAA-AA ....(((((((((.-----...).))).....((((.(((((((.........)))))))(.(((((((....))))))).).)))).......--...)))))........-.. ( -39.80) >DroSec_CAF1 127853 100 - 1 UAGCCCCCUGGCCG-----AAAGUGCCCGGGACACCUGUGUGUAUAUUAUUAUUACACAUCGGGCC--------AUGGGCGGCGGAGGGGGCCA--AAAAGGGGAUAAAAAACAA ..(((((((..(((-----....((((((((...((.(((((((.........))))))).)).))--------.)))))).))))))))))..--................... ( -41.30) >DroSim_CAF1 105597 100 - 1 UAGCCCCCUGGCCG-----AAAGUGCCCGGGACACCUGUGUGUAUAUUAUUAUUACACAUCGGGCC--------AUGGGCGGCGGAGGGGGCCA--AAAAGGGGAUAAAAAACAA ..(((((((..(((-----....((((((((...((.(((((((.........))))))).)).))--------.)))))).))))))))))..--................... ( -41.30) >DroEre_CAF1 114567 95 - 1 UAGCCCCCUGGCCG-----AAAGUGCCCGGGACACCUGUGUGUAUAUUAUUAUUACACAUCGGGCC--------AUGGGCGGCGGAGGG-GCCA--GAAGGGGGCUACA----AA (((((((((((((.-----...(((((((((...((.(((((((.........))))))).)).))--------.))))).)).....)-))).--..)))))))))..----.. ( -47.40) >DroYak_CAF1 118411 86 - 1 UAGCCCCCUGGCCG-----AAAGUGCCCGGGACACCUGUGUGUAUAUUAUUAUUACACAUUGGGCC--------AUGG-------AGGG-AUAAAAAAAGGGGG----A----GA ...((((((((((.-----...).)))..((...((.(((((((.........))))))).)).))--------....-------....-........))))))----.----.. ( -31.80) >DroPer_CAF1 112105 99 - 1 UAGCCCCCUGGCCGAACCGAAAGAGCCUGGGACACCUGUGUGUAUAUUAUUAUUACACAUCGGA-C--------AUGGCCAAGAGAACAGGGCA--AUGAG-----CAAUGGCAA ..((((((.(((.....(....).))).)))...((.(((((((.........))))))).)).-(--------((.(((..(....)..))).--)))..-----....))).. ( -29.70) >consensus UAGCCCCCUGGCCG_____AAAGUGCCCGGGACACCUGUGUGUAUAUUAUUAUUACACAUCGGGCC________AUGGGCGGCGGAGGGGGCCA__AAAAGGGGAUAAAAAA_AA ....((((((.(((..........((((((......((((((.(((....)))))))))))))))..........))).)(((.......)))......)))))........... (-20.69 = -22.22 + 1.53)

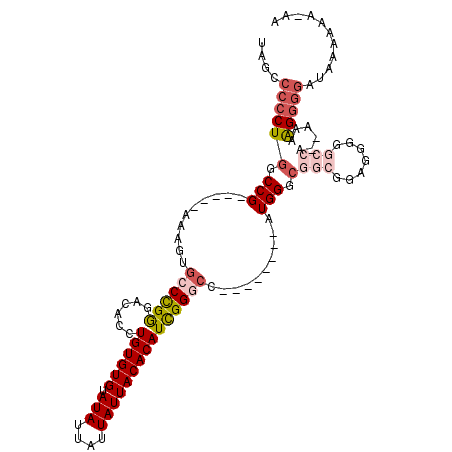
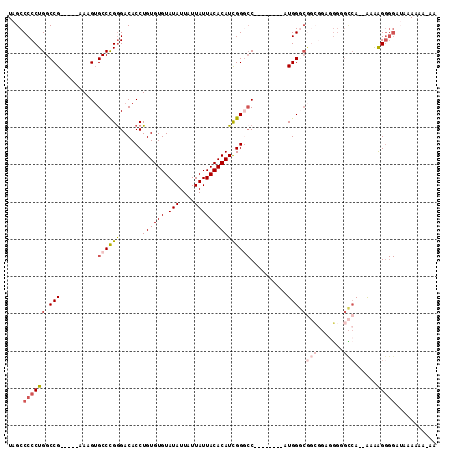
| Location | 15,321,966 – 15,322,056 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 82.66 |
| Mean single sequence MFE | -32.40 |
| Consensus MFE | -24.68 |
| Energy contribution | -23.92 |
| Covariance contribution | -0.76 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.929993 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15321966 90 + 20766785 UGCCAGGUUCGUACCUCAUCAAUAAGGUCCGCGAACGUGGAUGUGGAUG-GGAUACGAA------------------AGAUGGUUGGAUGGGUGAUGGG-GGGCGGGGCA .(((....(((..((((((((...(.((((((....)))))).).....-.....(...------------------.).............)))))))-)..)))))). ( -30.10) >DroSec_CAF1 127953 90 + 1 UGCCAGCUUCGUACCUCAUCAAUAAGGUCCGCGAACGUGGAUGUGGAUG-GGUUACAAA------------------AGAUGGU-GGUUGGGUGAUGGGGGGGCGGGGCA .....(((((((.((((((((...(.((((((....)))))).)...(.-((..((...------------------.....))-..)).).))))))))..))))))). ( -32.60) >DroSim_CAF1 105697 89 + 1 UGCCAGCUUCGUACCUCAUCAAUAAGGUCCGCGAACGUGGAUGUGGAUG-GGAUACGAA------------------AGAUGGU-GGAUGGGUGAUGGG-GGGCGGGGCA .....((((((..((((((((.....((((((...(((........)))-.....(...------------------.)...))-))))...)))))))-)..)))))). ( -33.70) >DroEre_CAF1 114662 93 + 1 CGCCAGCUUCGUACCUCAUCAAUAAGCUCCGCGAACGUGGACGUGGAUGUGGAUGUGGAUG---------------UGGAUGGA-AGAUGGUAGAUGGG-GGGCGGGGCA .....((((((..(((((((.((...(((((...((((..((((........))))..)))---------------)...))).-))...)).))))))-)..)))))). ( -32.70) >DroYak_CAF1 118497 108 + 1 UGCCAGCUUCGUACCUCAUCAAUAAGCUCCGCGAACGUGGAUAUGGAUGUGGAUGUGGAUGGAAGAUGGUAUAUGGUGGAUGGU-GGAUGGUGGAUGGG-GGGCGGGGCA .....((((((..(((((((.((..((((((((..(((....)))..)))))).))..)).....((.((.(((.(....).))-).)).)).))))))-)..)))))). ( -32.90) >consensus UGCCAGCUUCGUACCUCAUCAAUAAGGUCCGCGAACGUGGAUGUGGAUG_GGAUACGAA__________________AGAUGGU_GGAUGGGUGAUGGG_GGGCGGGGCA .....((((((..(((((((.......(((((....)))))((((........))))....................................)))))).)..)))))). (-24.68 = -23.92 + -0.76)

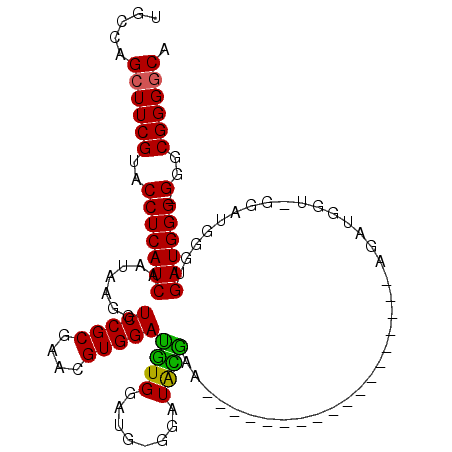

| Location | 15,322,027 – 15,322,136 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 80.08 |
| Mean single sequence MFE | -28.35 |
| Consensus MFE | -18.38 |
| Energy contribution | -18.60 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.704519 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15322027 109 + 20766785 UGGUUGGAUGGGUGAUGGGGGGCGG----------GGCAUUGGCGCUGCAUGUAGCACAUGUAGCGGCGAUUACGCGUAAUAAUCAUCAUAAUCAAAAGUCGCAACACGAAAUGAGUUU ..(((((((..(((((((...(((.----------(..((((.(((((((((.....))))))))).))))..).))).....)))))))........))).))))............. ( -38.00) >DroVir_CAF1 158518 86 + 1 -----------------------GG----------GGCGAUGGCGCUGCAUGUAGCACAUGUAGCGGCGAUUACGCGUAAUAAUCAUCAUAAUCAAAAGUCGCAACACGAAAUGAGUUU -----------------------(.----------.((((((.(((((((((.....))))))))).)(((((.......))))).............)))))..)............. ( -27.50) >DroGri_CAF1 131548 117 + 1 GGGG-GGGGGUGCGUUGGG-ACUGGGGCGGGUGAUGGCAAUGGCGCUGCAUGUAGCACAUGUAGCGGCGAUUACGCGUAAUAAUCAUCAUAAUCAAAAGUCGCAACACGAAAUGAGUUU ....-.....((.((((.(-(((.((...((((((.((.....(((((((((.....)))))))))(((....)))))....))))))....))...)))).)))).)).......... ( -38.40) >DroWil_CAF1 254617 86 + 1 -----------------------CG----------GGUAGUGGUGCUGCAUGUAGCACAUGUAGCGGCGAUUACGCGUAAUAAUCAUCAUAAUCAAAAGUCGCAACACGAAAUGAGUUU -----------------------((----------.(((((.((((((((((.....)))))))).)).))))).))...............(((....(((.....)))..))).... ( -24.00) >DroMoj_CAF1 161580 93 + 1 ---------------UGGC-AAUGG----------GGCGAUGGCGCUGCAUGUAGCACAUGUAUCAACGAUUACGCGUAAUAAUCAUCAUAAUCAAAAGUCGCAACACGAAAUGAGUUU ---------------....-..((.----------.(((((...(.((((((.....)))))).)...(((((.......))))).............)))))..))............ ( -17.40) >DroAna_CAF1 101724 93 + 1 ----------------AGGAGUGGG----------GGUACUGGCGCUGCAUGUAGCACAUGUAGCGGCGAUUACGCGUAAUAAUCAUCAUAAUCAAAAGUCGCAACACGAAAUGAGUUU ----------------....(((..----------((.((((.(((((((((.....))))))))).)(((((.......)))))............)))).)..)))........... ( -24.80) >consensus ________________GGG____GG__________GGCAAUGGCGCUGCAUGUAGCACAUGUAGCGGCGAUUACGCGUAAUAAUCAUCAUAAUCAAAAGUCGCAACACGAAAUGAGUUU .........................................(.(((((((((.....))))))))).)(((((.......))))).......(((....(((.....)))..))).... (-18.38 = -18.60 + 0.22)
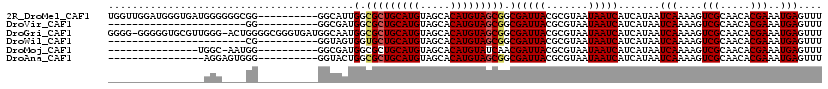
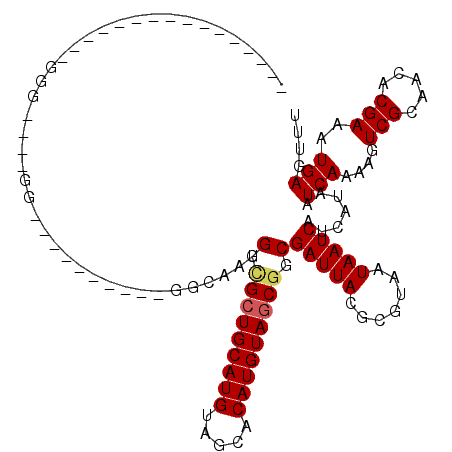
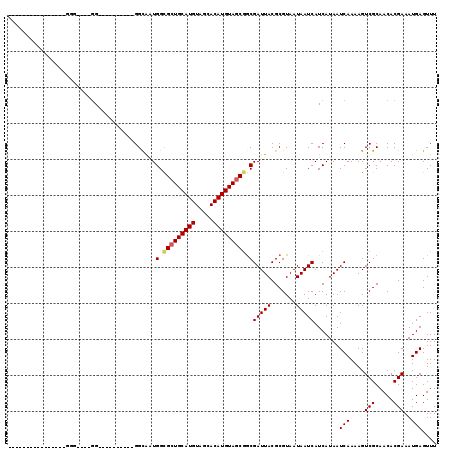
| Location | 15,322,056 – 15,322,151 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.63 |
| Mean single sequence MFE | -31.88 |
| Consensus MFE | -25.78 |
| Energy contribution | -25.42 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.826994 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15322056 95 + 20766785 UUGGCGCUGCAUGUAGCACAUGUAGCGGCGAUUACGCGUAAUAAUCAUCAUAAUCAAAAGUCGCAACACGAAAUGAGUUUCGACUUGCUG----------UUGCC--------------- ..(((((((((((.....))))))))((((((((.......)))))...........((((((.(((.........))).))))))))).----------..)))--------------- ( -29.00) >DroVir_CAF1 158524 100 + 1 AUGGCGCUGCAUGUAGCACAUGUAGCGGCGAUUACGCGUAAUAAUCAUCAUAAUCAAAAGUCGCAACACGAAAUGAGUUUUGACUUGUUG----------UUG------UUGCUGC---- ..(.(((((((((.....))))))))).)(((((.......))))).............((.((((((...((..(((....)))..)).----------.))------)))).))---- ( -31.00) >DroGri_CAF1 131585 110 + 1 AUGGCGCUGCAUGUAGCACAUGUAGCGGCGAUUACGCGUAAUAAUCAUCAUAAUCAAAAGUCGCAACACGAAAUGAGUUUUGACUUGUUG----------UUGUUGUUGUUUCGAGUGUU .((.(((((((((.....))))))))).))..(((.((.((((((.((.(((((...((((((.(((.........))).))))))))))----------).)).)))))).)).))).. ( -31.60) >DroEre_CAF1 114755 95 + 1 UUGGCGCUGCAUGUAGCACAUGUAGCGGCGAUUACGCGUAAUAAUCAUCAUAAUCAAAAGUCGCAACACGAAAUGAGUUUCGACUUGUUG----------UUGCC--------------- (((.(((((((((.....))))))))).)))......((((((((............((((((.(((.........))).))))))))))----------)))).--------------- ( -29.10) >DroWil_CAF1 254623 105 + 1 GUGGUGCUGCAUGUAGCACAUGUAGCGGCGAUUACGCGUAAUAAUCAUCAUAAUCAAAAGUCGCAACACGAAAUGAGUUUCGACUUGUUG----------UUGCUGUUGCGA-----GCU (((((((((((((.....)))))))).(((....))).........))))).........(((((((((((((..(((....)))..)).----------))).))))))))-----... ( -34.60) >DroAna_CAF1 101737 105 + 1 CUGGCGCUGCAUGUAGCACAUGUAGCGGCGAUUACGCGUAAUAAUCAUCAUAAUCAAAAGUCGCAACACGAAAUGAGUUUCGACUUGUUGUGAGCUGCCGUUGCU--------------- ..(((((((((((.....))))))))((((((((.......))))).(((((((...((((((.(((.........))).)))))))))))))))))))......--------------- ( -36.00) >consensus AUGGCGCUGCAUGUAGCACAUGUAGCGGCGAUUACGCGUAAUAAUCAUCAUAAUCAAAAGUCGCAACACGAAAUGAGUUUCGACUUGUUG__________UUGCU_______________ ..(.(((((((((.....))))))))).)(((((.......)))))...........((((((.(((.........))).)))))).................................. (-25.78 = -25.42 + -0.36)
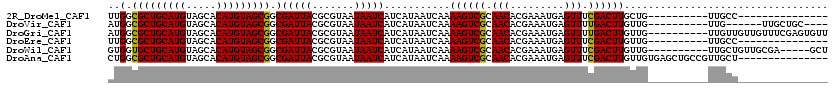
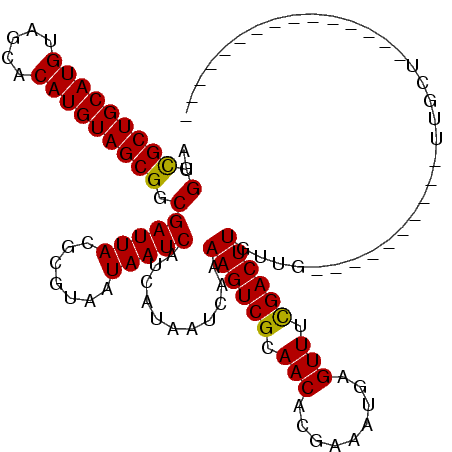
| Location | 15,322,056 – 15,322,151 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.63 |
| Mean single sequence MFE | -26.27 |
| Consensus MFE | -21.58 |
| Energy contribution | -22.08 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.612383 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15322056 95 - 20766785 ---------------GGCAA----------CAGCAAGUCGAAACUCAUUUCGUGUUGCGACUUUUGAUUAUGAUGAUUAUUACGCGUAAUCGCCGCUACAUGUGCUACAUGCAGCGCCAA ---------------(((..----------..((((((((.(((.(.....).))).))))))..(((((((.((.......))))))))))).(((.((((.....)))).)))))).. ( -29.20) >DroVir_CAF1 158524 100 - 1 ----GCAGCAA------CAA----------CAACAAGUCAAAACUCAUUUCGUGUUGCGACUUUUGAUUAUGAUGAUUAUUACGCGUAAUCGCCGCUACAUGUGCUACAUGCAGCGCCAU ----((.((((------((.----------.....(((....))).......)))))).......(((((((.((.......)))))))))))((((.((((.....)))).)))).... ( -24.12) >DroGri_CAF1 131585 110 - 1 AACACUCGAAACAACAACAA----------CAACAAGUCAAAACUCAUUUCGUGUUGCGACUUUUGAUUAUGAUGAUUAUUACGCGUAAUCGCCGCUACAUGUGCUACAUGCAGCGCCAU ....................----------....(((((..(((.(.....).)))..)))))..(((((((.((.......)))))))))(.((((.((((.....)))).)))).).. ( -21.00) >DroEre_CAF1 114755 95 - 1 ---------------GGCAA----------CAACAAGUCGAAACUCAUUUCGUGUUGCGACUUUUGAUUAUGAUGAUUAUUACGCGUAAUCGCCGCUACAUGUGCUACAUGCAGCGCCAA ---------------(((..----------....((((((.(((.(.....).))).))))))..(((((((.((.......))))))))))))(((.((((.....)))).)))..... ( -27.40) >DroWil_CAF1 254623 105 - 1 AGC-----UCGCAACAGCAA----------CAACAAGUCGAAACUCAUUUCGUGUUGCGACUUUUGAUUAUGAUGAUUAUUACGCGUAAUCGCCGCUACAUGUGCUACAUGCAGCACCAC .((-----((((((((((..----------......))(((((....))))))))))))).....(((((((.((.......))))))))))).(((.((((.....)))).)))..... ( -26.40) >DroAna_CAF1 101737 105 - 1 ---------------AGCAACGGCAGCUCACAACAAGUCGAAACUCAUUUCGUGUUGCGACUUUUGAUUAUGAUGAUUAUUACGCGUAAUCGCCGCUACAUGUGCUACAUGCAGCGCCAG ---------------.((...(((..........((((((.(((.(.....).))).))))))..(((((((.((.......))))))))))))(((.((((.....)))).)))))... ( -29.50) >consensus _______________AGCAA__________CAACAAGUCGAAACUCAUUUCGUGUUGCGACUUUUGAUUAUGAUGAUUAUUACGCGUAAUCGCCGCUACAUGUGCUACAUGCAGCGCCAA ..................................((((((.(((.(.....).))).))))))..(((((((.((.......)))))))))(.((((.((((.....)))).)))).).. (-21.58 = -22.08 + 0.50)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:26:38 2006