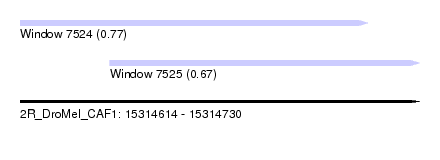
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 15,314,614 – 15,314,730 |
| Length | 116 |
| Max. P | 0.769713 |

| Location | 15,314,614 – 15,314,715 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.95 |
| Mean single sequence MFE | -31.22 |
| Consensus MFE | -19.19 |
| Energy contribution | -19.67 |
| Covariance contribution | 0.47 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.769713 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
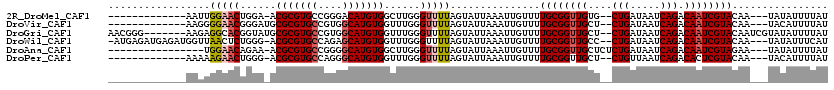
>2R_DroMel_CAF1 15314614 101 + 20766785 -------------AAUUGGAACUGGA-ACGCGUGCCGGGACAUGUGGCUUGGGUUUUAGUAUUAAAUUGUUUUGCGGUUGUG--CUGAUAAUCAGACAAUCGUACAA---UAUAUUUUAU -------------..((((.((((((-((.((.((((.......)))).)).)))))))).))))..((((.(((((((((.--(((.....)))))))))))).))---))........ ( -31.20) >DroVir_CAF1 147432 102 + 1 -------------AAGGGGAACGGGAUGCGCGUGCCGUGGCAUGUGGUUUGGGUUUUAGUAUUAAAUUGUUUUGCGGUUGCU--CUGAUAAUCAGACAAUCGUACAA---UACAUUUUAU -------------....(((((..((..(((((((....)))))))..))..))))).....((((.(((..((((((((.(--(((.....))))))))))))...---.))).)))). ( -29.60) >DroGri_CAF1 119552 111 + 1 AACGGG-------AAGAGGCACGGUAUGCGCGUGCCGUGGCAUGUGGUUUGGGUUUUAGUAUUAAAUUGUUUUGCGGUUGCU--CUGAUAAUCAGACAAUCGUACAAUCGUAUAUUUUAU .((((.-------....(.((((((((....)))))))).)..(..((((((.........))))))..)..((((((((.(--(((.....))))))))))))...))))......... ( -31.70) >DroWil_CAF1 237523 113 + 1 -AUGAGAUGAGAUGGUUAACUCUGGG-ACGCGUGCCAGAGCAUGUGGUUUGGGUUUUAGUAUUAAAUUGUUUUGCGGUUGCC--CUGAUAAUCAGACAAUCGUACAA---UAUAUUUCAU -((((((((.((((.(.(((((....-.(((((((....)))))))....)))))..).))))....((((.((((((((..--(((.....))).)))))))).))---)))))))))) ( -32.60) >DroAna_CAF1 93311 100 + 1 ----------------UGGAACAGAA-ACGCGUGCCGGGGCAUGUGGCUUGGGUUUUAGUAUUAAAUUGUUUUGCGGUUGCUCUCUGAUAAUCAGACAAUCGUAGAA---UAUAUUUUAU ----------------((((((.((.-.(((((((....)))))))..))..))))))....((((.(((((((((((((...((((.....)))))))))))))))---))...)))). ( -32.90) >DroPer_CAF1 104978 101 + 1 -------------AAAAAGAACUGGG-ACGCGUGCCAGGGCAUGUGGUUUGGGUUUUAGUAUUAAAUUGUUUUGCGGUUGCU--CUGUUAAUCAGACACUCGUACAA---UACAUUUUAU -------------....((((((.((-((((((((....)))))).)))).)))))).....((((.(((..((((..((.(--(((.....))))))..))))...---.))).)))). ( -29.30) >consensus _____________AAGUGGAACUGGA_ACGCGUGCCGGGGCAUGUGGUUUGGGUUUUAGUAUUAAAUUGUUUUGCGGUUGCU__CUGAUAAUCAGACAAUCGUACAA___UAUAUUUUAU .................(((((......(((((((....)))))))......)))))...............((((((((....(((.....))).))))))))................ (-19.19 = -19.67 + 0.47)
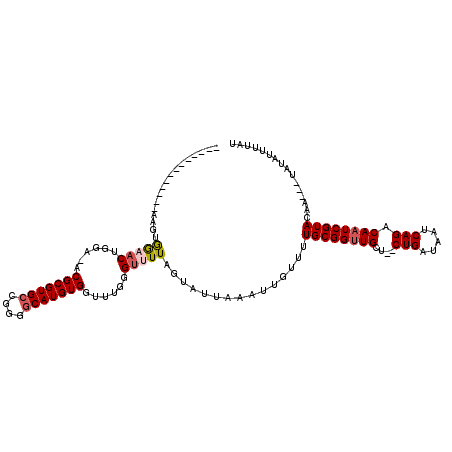
| Location | 15,314,640 – 15,314,730 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.86 |
| Mean single sequence MFE | -24.94 |
| Consensus MFE | -11.09 |
| Energy contribution | -12.03 |
| Covariance contribution | 0.95 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.44 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.672194 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15314640 90 + 20766785 CAUGUGGCUUGGGUUUUAGUAUUAAAUUGUUUUGCGGUUGUG--CUGAUAAUCAGACAAUCGUACAA---UAUAUUUUAUUGGAGUGGC---------UU-----CCC----A------- ...((.((((.(((...(((((...(((((...((((((((.--(((.....)))))))))))))))---))))))..))).)))).))---------..-----...----.------- ( -24.60) >DroGri_CAF1 119585 104 + 1 CAUGUGGUUUGGGUUUUAGUAUUAAAUUGUUUUGCGGUUGCU--CUGAUAAUCAGACAAUCGUACAAUCGUAUAUUUUAUUGAAGUGCC---------CA-----ACUGCCCACUCCUAU ...((((.(((((((((((((.((.((.(((.((((((((.(--(((.....)))))))))))).))).)).))...)))))))..)))---------))-----)....))))...... ( -30.10) >DroWil_CAF1 237561 106 + 1 CAUGUGGUUUGGGUUUUAGUAUUAAAUUGUUUUGCGGUUGCC--CUGAUAAUCAGACAAUCGUACAA---UAUAUUUCAUUGGACUGGAGUGGUUUUCUU-----CCC----AUUCGUUU .....((((..(.....(((((...(((((...(((((((..--(((.....))).)))))))))))---))))))...)..)))).((((((.......-----.))----)))).... ( -25.00) >DroMoj_CAF1 146994 84 + 1 CAUGUGGUUUGGGUUUUAGUAUUAAAUUGUUUUGCGGUUGCU--CUGAUAAUCAGACAAUCGUACAA---UAUAUUUUAUUGC--UGCC---------UC-------------------- ..........((((...((((.((((.((((.((((((((.(--(((.....)))))))))))).))---))...)))).)))--))))---------).-------------------- ( -23.20) >DroAna_CAF1 93334 92 + 1 CAUGUGGCUUGGGUUUUAGUAUUAAAUUGUUUUGCGGUUGCUCUCUGAUAAUCAGACAAUCGUAGAA---UAUAUUUUAUUGGAGUGGA---------CU-----CGA----G------- .......(((((((((......((((.(((((((((((((...((((.....)))))))))))))))---))...)))).......)))---------))-----)))----)------- ( -26.12) >DroPer_CAF1 105004 95 + 1 CAUGUGGUUUGGGUUUUAGUAUUAAAUUGUUUUGCGGUUGCU--CUGUUAAUCAGACACUCGUACAA---UACAUUUUAUUGGAGUGGC---------UCUGCCUCUG----A------- ((.(.(((..(((((.......((((.(((..((((..((.(--(((.....))))))..))))...---.))).)))).......)))---------)).))).)))----.------- ( -20.64) >consensus CAUGUGGUUUGGGUUUUAGUAUUAAAUUGUUUUGCGGUUGCU__CUGAUAAUCAGACAAUCGUACAA___UAUAUUUUAUUGGAGUGGC_________UU_____CCC____A_______ ......((((.(((...(((((....((((...(((((((....(((.....))).)))))))))))....)))))..))).)))).................................. (-11.09 = -12.03 + 0.95)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:26:31 2006