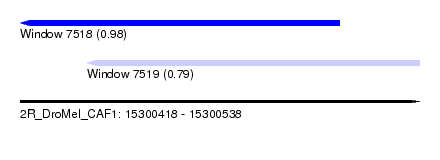
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 15,300,418 – 15,300,538 |
| Length | 120 |
| Max. P | 0.981065 |

| Location | 15,300,418 – 15,300,514 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 75.22 |
| Mean single sequence MFE | -24.00 |
| Consensus MFE | -17.19 |
| Energy contribution | -16.92 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.88 |
| SVM RNA-class probability | 0.981065 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
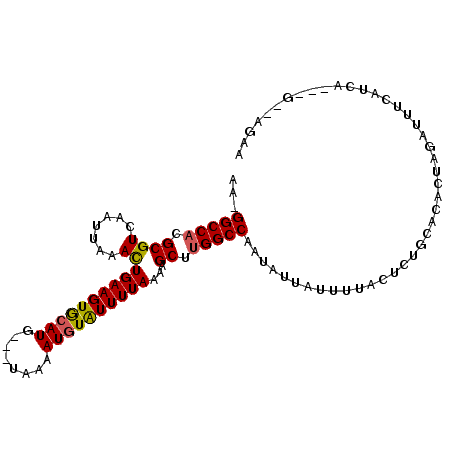
>2R_DroMel_CAF1 15300418 96 - 20766785 AA-GGCCACGCGUCAAUUAUACUGAAGUAGAUG---UAAAAUGUAUUUUAAAAGCUUGGCCAAUAUUAUUUUACCAUGCGCACUGGAUUUCAUCG---G--GGCA ..-(((((.((((.......))(((((((.((.---....)).)))))))...)).)))))..................((.((.(((...))).---)--))). ( -20.90) >DroVir_CAF1 123881 98 - 1 GA-GGCCACGCGUCAAUUAAACUGAAGUGCAUG---UAAAAUGUAUUUUAAAAGCUUGGCCAAUAUUAUUUUACUCGCCAA-GUAUAUUUGAUUCGUGC--GGAA ..-.((.(((.(((((.((...((((((((((.---....))))))))))...(((((((...((......))...)))))-)).)).))))).)))))--.... ( -27.00) >DroPse_CAF1 131714 99 - 1 AAGGGCCACGCGUCAAUUAGACUGAAGUGCAUG---UAAAAUGUGUUUUAAAAGCCGGGCCAAUAUUAUCUUACUUUGCACUCUGGAUGGUAUCA---GCCAACA ...(((.....(((.....)))(((((..(((.---....)))..)))))...))).(((..(((((((((.............)))))))))..---))).... ( -25.32) >DroWil_CAF1 205988 88 - 1 AA-GGCCACGCGUCAAUUAAAUUGAAGUGCAUG---UAAAAUGUAUUUUAAAAGCUUGGCCAAUAUUGUAUUACUGUGAGCACA--------UCA---G--AGAA ..-(((((.((..........(((((((((((.---....)))))))))))..)).)))))............((.(((.....--------)))---.--)).. ( -23.70) >DroMoj_CAF1 122627 98 - 1 GA-GGCCACGCGUCAAUUAAAUUGAAGUGCAUG---UAAAAUGUAUUUUAAAAGCUUGGCCAAUAUUAUUUUACUCUCCAA--UACAUUUGAUUCGUGU-AAGAA ..-(((((.((..........(((((((((((.---....)))))))))))..)).))))).......((((((.(..(((--.....)))....).))-)))). ( -22.30) >DroAna_CAF1 78970 96 - 1 AG-GGCCACGCGUCAAUUAGAUUGAAGUACAUGGUAAAAAAUGUAUUUUAAAAGCUUGGCCAAUAUUGCUUCACU------ACUAGAUUUCAUCGACGG--GGCA ..-(((((.((.((.....))(((((((((((........)))))))))))..)).))))).....((((((...------................))--)))) ( -24.81) >consensus AA_GGCCACGCGUCAAUUAAACUGAAGUGCAUG___UAAAAUGUAUUUUAAAAGCUUGGCCAAUAUUAUUUUACUCUGCACACUAGAUUUCAUCA___G__AGAA ...(((((.((((.......))((((((((((........))))))))))...)).)))))............................................ (-17.19 = -16.92 + -0.28)

| Location | 15,300,438 – 15,300,538 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 75.87 |
| Mean single sequence MFE | -22.14 |
| Consensus MFE | -16.36 |
| Energy contribution | -16.28 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.09 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.787234 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
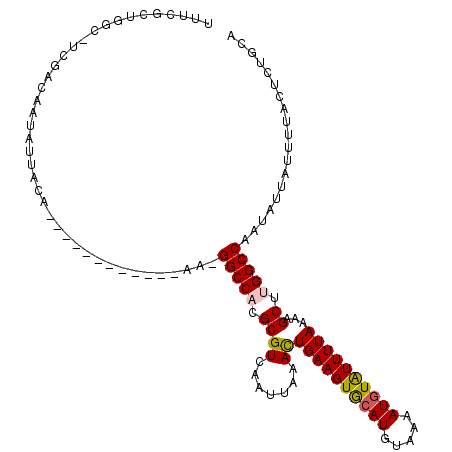
>2R_DroMel_CAF1 15300438 100 - 20766785 UUUCCUUGUC-UCGACUAUAUCACA------------AA-GGCCACGCGUCAAUUAUACUGAAGUAGAUGUAAAAUGUAUUUUAAAAGCUUGGCCAAUAUUAUUUUACCAUGCG ..........-..............------------..-(((((.((((.......))(((((((.((.....)).)))))))...)).)))))................... ( -15.60) >DroVir_CAF1 123903 109 - 1 UUUCGCCUGC-CUUUCAAUAUAACAGU---GGAACACGA-GGCCACGCGUCAAUUAAACUGAAGUGCAUGUAAAAUGUAUUUUAAAAGCUUGGCCAAUAUUAUUUUACUCGCCA ..(((....(-(...............---))....)))-(((((.((((.......))((((((((((.....))))))))))...)).)))))................... ( -22.16) >DroPse_CAF1 131736 100 - 1 UUUCGCUGCCUUCGAG--UAUUACA------------AAGGGCCACGCGUCAAUUAGACUGAAGUGCAUGUAAAAUGUGUUUUAAAAGCCGGGCCAAUAUUAUCUUACUUUGCA .......((....(((--((.....------------...((((..(((((.....)))(((((..(((.....)))..)))))...))..))))..........))))).)). ( -22.87) >DroWil_CAF1 206000 112 - 1 UUUCACUUGC-CUUCCAAUAUUACAAAAAACAGAAAAAA-GGCCACGCGUCAAUUAAAUUGAAGUGCAUGUAAAAUGUAUUUUAAAAGCUUGGCCAAUAUUGUAUUACUGUGAG ..((((....-....(((((((.(........)......-(((((.((..........(((((((((((.....)))))))))))..)).)))))))))))).......)))). ( -24.86) >DroMoj_CAF1 122649 99 - 1 UUUCGCUCGA----GCAAUA----------GGAAGACGA-GGCCACGCGUCAAUUAAAUUGAAGUGCAUGUAAAAUGUAUUUUAAAAGCUUGGCCAAUAUUAUUUUACUCUCCA ....((.(((----((....----------....((((.-(....).)))).......(((((((((((.....)))))))))))..))))))).................... ( -24.50) >DroPer_CAF1 90362 100 - 1 UUUCGCUGCCUUCGAG--UAUUACA------------AAGGGCCACGCGUCAAUUAGACUGAAGUGCAUGUAAAAUGUGUUUUAAAAGCCGGGCCAAUAUUAUCUUACUUUGCA .......((....(((--((.....------------...((((..(((((.....)))(((((..(((.....)))..)))))...))..))))..........))))).)). ( -22.87) >consensus UUUCGCUGGC_UCGACAAUAUUACA____________AA_GGCCACGCGUCAAUUAAACUGAAGUGCAUGUAAAAUGUAUUUUAAAAGCUUGGCCAAUAUUAUUUUACUCUGCA ........................................(((((.((((.......))((((((((((.....))))))))))...)).)))))................... (-16.36 = -16.28 + -0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:26:24 2006