| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 15,286,947 – 15,287,037 |
| Length | 90 |
| Max. P | 0.856006 |

| Location | 15,286,947 – 15,287,037 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 80.28 |
| Mean single sequence MFE | -22.12 |
| Consensus MFE | -16.45 |
| Energy contribution | -16.45 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.856006 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
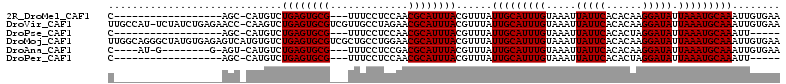
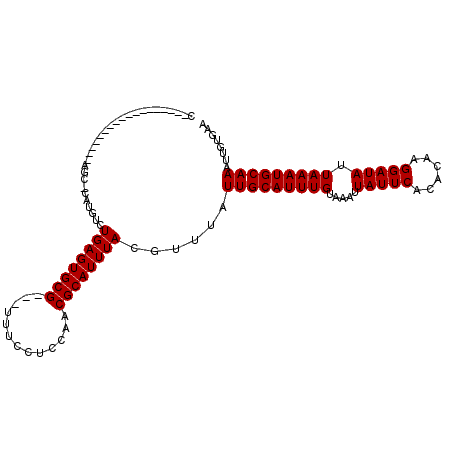
>2R_DroMel_CAF1 15286947 90 - 20766785 C------------------AGC-CAUGUCUGAGUGCG---UUUCCUCCAACGCAUUUACGUUUAUUGCAUUUGUAAAUUAUUCACACAAGGAUAUUAAAUGCAAAUUGUGAA .------------------...-(((...((((((((---((......))))))))))......(((((((((.....(((((......))))).)))))))))...))).. ( -21.10) >DroVir_CAF1 107202 110 - 1 UUGCCAU-UCUAUCUGAGAACC-CAAGUCUGAGUGCGUCGUUGCCUAGAACGCAUUUACGUUUAUUGCAUUUGUAAAUUAUUCACACAAGGAUAUUAAAUGCAAAUUGUGAA ......(-(((.....))))..-.(((..((((((((((........).)))))))))..))).(((((((((.....(((((......))))).)))))))))........ ( -21.60) >DroPse_CAF1 120183 85 - 1 C------------------AGC-CAUGUCUGAGUGCG---UUUCCUCCAACGCAUUUACGUUUAUUGCAUUUGUAAAUUAUUCACACUAGGAUAUUAAAUGCAAAUU----- .------------------...-......((((((((---((......))))))))))......(((((((((.....(((((......))))).)))))))))...----- ( -20.40) >DroMoj_CAF1 106294 112 - 1 UUGGCAGGGCUAUGUGAGAGUCAUGUGUCUGAGUGCGUCGCUGCCUGGAACGCAUUUACGUUUAUUGCAUUUGUAAAUUAUUCACACAAGGAUAUUAAAUGCAAAUUGUGAA ..(((((((((((...(((........)))..))).))).)))))..(((((......))))).(((((((((.....(((((......))))).)))))))))........ ( -26.20) >DroAna_CAF1 68860 94 - 1 C----AU-G--------G-AGU-CAUGUCUGAGUGCG---UUUCCUCCGACGCAUUUACGUUUAUUGCAUUUGUAAAUUAUUCACACAAGGAUAUUAAAUGCAAAUUGUGAA .----..-.--------.-..(-(((...((((((((---((......))))))))))......(((((((((.....(((((......))))).)))))))))...)))). ( -23.00) >DroPer_CAF1 78769 85 - 1 C------------------AGC-CAUGUCUGAGUGCG---UUUCCUCCAACGCAUUUACGUUUAUUGCAUUUGUAAAUUAUUCACACUAGGAUAUUAAAUGCAAAUU----- .------------------...-......((((((((---((......))))))))))......(((((((((.....(((((......))))).)))))))))...----- ( -20.40) >consensus C__________________AGC_CAUGUCUGAGUGCG___UUUCCUCCAACGCAUUUACGUUUAUUGCAUUUGUAAAUUAUUCACACAAGGAUAUUAAAUGCAAAUUGUGAA .............................((((((((.............))))))))......(((((((((.....(((((......))))).)))))))))........ (-16.45 = -16.45 + 0.00)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:26:22 2006