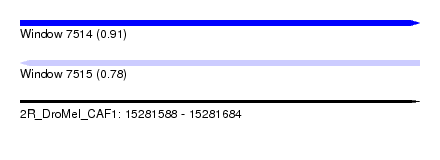
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 15,281,588 – 15,281,684 |
| Length | 96 |
| Max. P | 0.911731 |

| Location | 15,281,588 – 15,281,684 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.99 |
| Mean single sequence MFE | -23.75 |
| Consensus MFE | -16.80 |
| Energy contribution | -16.72 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.911731 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
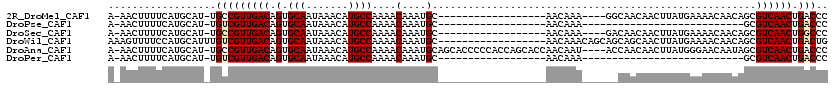
>2R_DroMel_CAF1 15281588 96 + 20766785 GGGUCAGUUGACGCUGUUGUUUUCAUAAGUUGUUGCC----UUUGUU------------------GCAUUUGUUUUGGCAUGUUUAUUGCACUGUCAACGGCA-AUGCAUGAAAAGUU-U (.(((....))).).....(((((((..(((((((..----......------------------((....)).(((((((((.....))).)))))))))))-))..)))))))...-. ( -20.80) >DroPse_CAF1 112616 73 + 1 GGGUCAGUUGACGC---------------------------UUUGUU------------------GCAUUUGUUUUGGCAUGUUUAUUGCACUGUCAACGACA-AUGCAUGAAAAGUU-U ..(((....)))((---------------------------(((..(------------------(((((.((((((((((((.....))).)))))).))))-)))))...))))).-. ( -21.30) >DroSec_CAF1 90932 96 + 1 GGGCCAGUUGACGCUGUUGUUUUCAUAAGUUGUUGUC----UUUGUU------------------GCAUUUGUUUUGGCAUGUUUAUUGCACUGUCAACGGCA-AUGCAUGAAAAGUU-U ....((((....))))...(((((((..((((((((.----......------------------((....))..((((((((.....))).)))))))))))-))..)))))))...-. ( -19.80) >DroWil_CAF1 171622 102 + 1 CAGUCAGUUGACGCUGUUGUUUUCAUAAGUUGCUGCUGCUGUUUGUU------------------GCAUUUGUUUUGGCAUGUUUAUUGCACUGUCAACGACAAAUGCAUGGAAAACUUU ..(((....)))......(((((((((((..((....))..)))))(------------------((((((((((((((((((.....))).)))))).))))))))))..))))))... ( -30.70) >DroAna_CAF1 63665 114 + 1 GGGUCAGUUGACGCUAUUGUUCCCAUAAGUUGUUGGU----AUUGUUGGUGCUGGUGGGGGUGCUGCAUUUGUUUUGGCAUGUUUAUUGCACUGUCAACGGCA-AUGCAUGAAAAGUU-U (((.((((.......))))..)))....((((((((.----.....((((((.((((((.(((((((....))...))))).)))))))))))))))))))).-..............-. ( -28.60) >DroPer_CAF1 71147 73 + 1 GGGUCAGUUGACGC---------------------------UUUGUU------------------GCAUUUGUUUUGGCAUGUUUAUUGCACUGUCAACGACA-AUGCAUGAAAAGUU-U ..(((....)))((---------------------------(((..(------------------(((((.((((((((((((.....))).)))))).))))-)))))...))))).-. ( -21.30) >consensus GGGUCAGUUGACGCUGUUGUUUUCAUAAGUUGUUG______UUUGUU__________________GCAUUUGUUUUGGCAUGUUUAUUGCACUGUCAACGACA_AUGCAUGAAAAGUU_U ..(((....))).....................................................((((.(((((((((((((.....))).)))))).)))).))))............ (-16.80 = -16.72 + -0.08)



| Location | 15,281,588 – 15,281,684 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.99 |
| Mean single sequence MFE | -16.08 |
| Consensus MFE | -8.32 |
| Energy contribution | -8.48 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.784412 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15281588 96 - 20766785 A-AACUUUUCAUGCAU-UGCCGUUGACAGUGCAAUAAACAUGCCAAAACAAAUGC------------------AACAAA----GGCAACAACUUAUGAAAACAACAGCGUCAACUGACCC .-...(((((((((((-((.......))))))........((((...........------------------......----)))).......))))))).......(((....))).. ( -16.43) >DroPse_CAF1 112616 73 - 1 A-AACUUUUCAUGCAU-UGUCGUUGACAGUGCAAUAAACAUGCCAAAACAAAUGC------------------AACAAA---------------------------GCGUCAACUGACCC .-.........(((((-((((...)))))))))....................((------------------......---------------------------))(((....))).. ( -14.30) >DroSec_CAF1 90932 96 - 1 A-AACUUUUCAUGCAU-UGCCGUUGACAGUGCAAUAAACAUGCCAAAACAAAUGC------------------AACAAA----GACAACAACUUAUGAAAACAACAGCGUCAACUGGCCC .-..............-.(((((((((.(.(((.......)))).........((------------------....((----(.......))).((....))...)))))))).))).. ( -13.90) >DroWil_CAF1 171622 102 - 1 AAAGUUUUCCAUGCAUUUGUCGUUGACAGUGCAAUAAACAUGCCAAAACAAAUGC------------------AACAAACAGCAGCAGCAACUUAUGAAAACAACAGCGUCAACUGACUG ...((((((..(((((((((..(((.((((.......)).)).))).))))))))------------------).......((....)).......))))))......(((....))).. ( -21.90) >DroAna_CAF1 63665 114 - 1 A-AACUUUUCAUGCAU-UGCCGUUGACAGUGCAAUAAACAUGCCAAAACAAAUGCAGCACCCCCACCAGCACCAACAAU----ACCAACAACUUAUGGGAACAAUAGCGUCAACUGACCC .-..............-....((((((.((((........(((..........))))))).((((..((..........----........))..)))).........))))))...... ( -15.67) >DroPer_CAF1 71147 73 - 1 A-AACUUUUCAUGCAU-UGUCGUUGACAGUGCAAUAAACAUGCCAAAACAAAUGC------------------AACAAA---------------------------GCGUCAACUGACCC .-.........(((((-((((...)))))))))....................((------------------......---------------------------))(((....))).. ( -14.30) >consensus A_AACUUUUCAUGCAU_UGCCGUUGACAGUGCAAUAAACAUGCCAAAACAAAUGC__________________AACAAA______CAACAACUUAUGAAAACAACAGCGUCAACUGACCC ..................(((((((((.(.(((.......))))....(....)......................................................)))))).))).. ( -8.32 = -8.48 + 0.17)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:26:20 2006