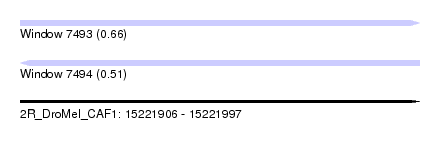
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 15,221,906 – 15,221,997 |
| Length | 91 |
| Max. P | 0.655929 |

| Location | 15,221,906 – 15,221,997 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 80.25 |
| Mean single sequence MFE | -16.00 |
| Consensus MFE | -8.72 |
| Energy contribution | -8.42 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.31 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.655929 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
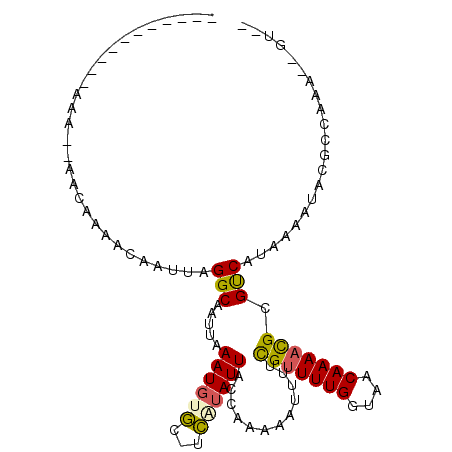
>2R_DroMel_CAF1 15221906 91 + 20766785 -----------AAA--AACAAAACAAUUAGGCAAUUAAAUGUCU-UGGUAUUACCAAAAAUUUUCGUUUUGGUAACAAAACGCGUCAUAAAAUAAGCCAAAAUGU-- -----------...--......(((.((.(((...........(-(((.....)))).......(((((((....))))))).............))).)).)))-- ( -15.90) >DroVir_CAF1 77938 89 + 1 -----------AAA--AACAAAACAAUUAGGCAAUUAAAUGUGC-UCAUAUUACCAAAAAUUUUCGUUUUGGUAACAAAACGCGUCAUAAAAUACGGCCAA--GG-- -----------...--.............(((.(((..((((((-.....(((((((((.......)))))))))......))).)))..)))...)))..--..-- ( -15.30) >DroGri_CAF1 61738 84 + 1 -----------AAGGAAACAAAACAAUUAGGCAAUUAAAUUCGC-UCAUAUUACCAAAAAUUUUCGUUUUGGUAACAAAACGCGUCGUAAAAGGCC----------- -----------..(....)..........(((..(((....(((-.....(((((((((.......)))))))))......)))...)))...)))----------- ( -16.20) >DroWil_CAF1 132734 105 + 1 CAAACGAAAAACAA--AACAAAACAAUUAGGCAAUUAAAUGUGACUCUUAUUACCAAAAAUUUUCGCUUUGGUAACAAAAUGCGCCAUAAAAUAGGCCAAAAAGUGU ..............--......(((.((((....)))).))).((.(((.((((((((.........)))))))).....((.(((........)))))..))).)) ( -13.30) >DroMoj_CAF1 74774 89 + 1 -----------AAG--AACAAAACAAUUAGGCAAUUAAAUGUGC-GCAUAUUACCGAAAAUUUUUGUUUUGGUAACAAAACGCGUCAUAAAAUGCCGCCAA--GG-- -----------...--.............((((.....((((((-(....(((((((((.......))))))))).....)))).)))....)))).....--..-- ( -18.50) >DroPer_CAF1 56636 92 + 1 -----------AAA--AACAAAACAAUUAGGCAAUUAAAUGUCUUUGGUAUUACCAAAAAUUUUCGUUUUGGUAACAAAACGCGUCAUAAAAUAAGCCAAAAUGU-- -----------...--......(((.((.(((...........(((((.....)))))......(((((((....))))))).............))).)).)))-- ( -16.80) >consensus ___________AAA__AACAAAACAAUUAGGCAAUUAAAUGUGC_UCAUAUUACCAAAAAUUUUCGUUUUGGUAACAAAACGCGUCAUAAAAUACGCCAAA__GU__ .............................(((.....((((((...))))))............(((((((....))))))).)))..................... ( -8.72 = -8.42 + -0.30)


| Location | 15,221,906 – 15,221,997 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 80.25 |
| Mean single sequence MFE | -19.62 |
| Consensus MFE | -9.63 |
| Energy contribution | -9.86 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.49 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.507438 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
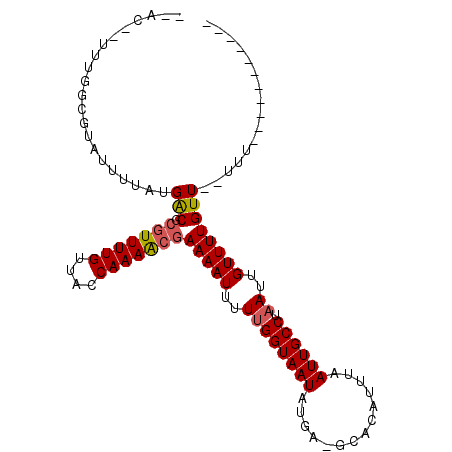
>2R_DroMel_CAF1 15221906 91 - 20766785 --ACAUUUUGGCUUAUUUUAUGACGCGUUUUGUUACCAAAACGAAAAUUUUUGGUAAUACCA-AGACAUUUAAUUGCCUAAUUGUUUUGUU--UUU----------- --(((.((.(((..(((..(((...(((((((....))))))).....((((((.....)))-))))))..))).))).)).)))......--...----------- ( -19.10) >DroVir_CAF1 77938 89 - 1 --CC--UUGGCCGUAUUUUAUGACGCGUUUUGUUACCAAAACGAAAAUUUUUGGUAAUAUGA-GCACAUUUAAUUGCCUAAUUGUUUUGUU--UUU----------- --..--..(((...(((..(((..((....(((((((((((.......)))))))))))...-)).)))..))).))).............--...----------- ( -18.10) >DroGri_CAF1 61738 84 - 1 -----------GGCCUUUUACGACGCGUUUUGUUACCAAAACGAAAAUUUUUGGUAAUAUGA-GCGAAUUUAAUUGCCUAAUUGUUUUGUUUCCUU----------- -----------(((..((.....(((....(((((((((((.......)))))))))))...-))).....))..)))..................----------- ( -18.70) >DroWil_CAF1 132734 105 - 1 ACACUUUUUGGCCUAUUUUAUGGCGCAUUUUGUUACCAAAGCGAAAAUUUUUGGUAAUAAGAGUCACAUUUAAUUGCCUAAUUGUUUUGUU--UUGUUUUUCGUUUG ..((..((.(((..(((..(((..((.((((((((((((((.......))))))))))))))))..)))..))).))).))..))......--.............. ( -21.90) >DroMoj_CAF1 74774 89 - 1 --CC--UUGGCGGCAUUUUAUGACGCGUUUUGUUACCAAAACAAAAAUUUUCGGUAAUAUGC-GCACAUUUAAUUGCCUAAUUGUUUUGUU--CUU----------- --..--.....((((((..(((..((((..(((((((((((......)))).))))))).))-)).)))..)).)))).............--...----------- ( -19.80) >DroPer_CAF1 56636 92 - 1 --ACAUUUUGGCUUAUUUUAUGACGCGUUUUGUUACCAAAACGAAAAUUUUUGGUAAUACCAAAGACAUUUAAUUGCCUAAUUGUUUUGUU--UUU----------- --(((.((.(((..(((..(((...(((((((....)))))))....(((((((.....))))))))))..))).))).)).)))......--...----------- ( -20.10) >consensus __AC__UUUGGCGUAUUUUAUGACGCGUUUUGUUACCAAAACGAAAAUUUUUGGUAAUAUGA_GCACAUUUAAUUGCCUAAUUGUUUUGUU__UUU___________ .....................(((.(((((((....)))))))(((((..((((((((..............)))))).))..))))))))................ ( -9.63 = -9.86 + 0.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:25:59 2006