| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 15,209,964 – 15,210,065 |
| Length | 101 |
| Max. P | 0.842274 |

| Location | 15,209,964 – 15,210,065 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 79.49 |
| Mean single sequence MFE | -25.32 |
| Consensus MFE | -16.94 |
| Energy contribution | -16.58 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.842274 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
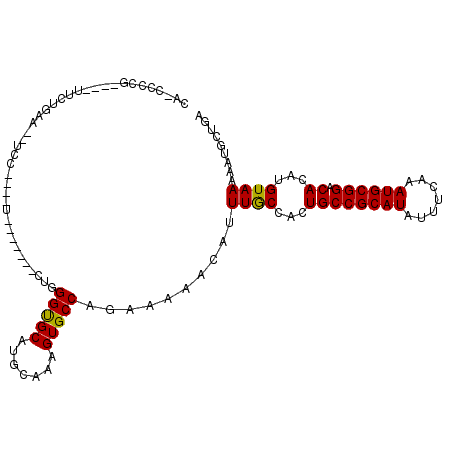
>2R_DroMel_CAF1 15209964 101 + 20766785 CA-CCCCG----UUCUGCAUUUCCUUCAU-------AUGGGUGCAUGCAAAGUGCCAGAAAAACAUUUACCAUUGCCGCAUAUUUCAAAUGCGGACACAUGUAAAAAUGCUGA ..-.....----.((.((((((.....((-------(((((..(.......)..)).................((((((((.......)))))).)))))))..)))))).)) ( -22.10) >DroPse_CAF1 73825 93 + 1 GAGCCCGG----AUCCGAC---------U-------CUGGGUGCAUGCAAUGUGCCAGAAAAACAGUUGCCACUGCCGCAUAUUUCAAAUGCGGACACAUGUAAAAAUGCUAA ..((((((----(......---------)-------))))))((((...(((((.........((((....))))((((((.......)))))).)))))......))))... ( -28.90) >DroEre_CAF1 40553 101 + 1 CA-CCCCA----UUCUGCAUUUCCCUCAU-------ACGGGUGCAUGCAAAGUGCCAGAAAAACAUUUACCACUGCCGCAUAUUUCAAAUGCGGACACAUGUAAAAAUGCUGA ..-...((----((.(((((..(((....-------..)))...))))).)))).(((.......(((((...((((((((.......)))))).))...)))))....))). ( -25.20) >DroWil_CAF1 101332 112 + 1 AA-CCCUGCAAAUUCUGCAAUUGCUGCAACUCGACGCUGGGCGCAUGCAAAGUGCCAGAAAAACAUUUGCCAUUGCCGCAUAUUUCAAAUGCGGACACAUGUAAAAAUGUUGA ..-.........(((((((.(((((((..((((....)))).))).))))..)).))))).(((((((..(((((((((((.......)))))).)).)))...))))))).. ( -30.10) >DroAna_CAF1 35419 87 + 1 ------------UGCUAAAG-UCC-------------UGGGUGCAUGCAAAGUGCCAGAAAAACAUUUGCCAUUGCCGCAUAUUUCAAAUGCGGACACAUGUAAAAAUGCUGA ------------.((.....-..(-------------((.(..(.......)..)))).......(((((...((((((((.......)))))).))...)))))...))... ( -18.60) >DroPer_CAF1 44777 93 + 1 GAGCCCGG----AUCCGAC---------U-------CUGGGUGCAUGCAAAGUGCCAGAAAAACAGUUGCCACUGCCGCAUAUUUCAAAUGCGGACACAUGUAAAAAUGCUGA ..((((((----(......---------)-------))))))(((((...((((.(((........))).)))).((((((.......))))))...)))))........... ( -27.00) >consensus CA_CCCCG____UUCUGAA__UCC____U_______CUGGGUGCAUGCAAAGUGCCAGAAAAACAUUUGCCACUGCCGCAUAUUUCAAAUGCGGACACAUGUAAAAAUGCUGA .......................................(((((.......)))))..........((((...((((((((.......)))))).))...))))......... (-16.94 = -16.58 + -0.36)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:25:57 2006