| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 15,209,872 – 15,209,962 |
| Length | 90 |
| Max. P | 0.755310 |

| Location | 15,209,872 – 15,209,962 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 74.06 |
| Mean single sequence MFE | -15.53 |
| Consensus MFE | -10.48 |
| Energy contribution | -10.65 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.18 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.755310 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15209872 90 + 20766785 UCGUUAUAAUAAAUAACUUCAACUGUCCG-ACACUUGACACUUGACAUCAGCUGCAGCGGAAGCGGCAACA------GGAGCCGCAAAAAGAA------AAUC--------- ..(((((.....))))).((((.((((..-......)))).)))).......(((.((....))(((....------...)))))).......------....--------- ( -20.20) >DroVir_CAF1 59700 94 + 1 UCGUUAUAAUAAAUAACUUCAACUGUCCA-GCACUUGACACUUGACAUCAACUGCAACGGAAGUAGCA-CA-------------CAAAAAAGUAC---GAGUCCGAACAAUU ..(((((.....))))).((((.((((..-......)))).)))).......(((.((....)).)))-..-------------.......((.(---(....)).)).... ( -15.30) >DroGri_CAF1 44461 97 + 1 UCGUUAUAAUAAAUAACUUCAACUGUCCA-ACACUUGACACUUGACAUCAACUGCAACGGAAGCCACA-CA-------------CAAAAAGCAAACGAGAGAAUAAACUAUU (((((.............((((.((((..-......)))).))))............(....).....-..-------------.........))))).............. ( -10.40) >DroWil_CAF1 101239 91 + 1 UCGUUAUAAUAAAUAACAUCAACUGUCCAAACACUUGACACUUGACAUCAACUGCAACGGAAGCGGCAAAA------CGUCACCCAAAACCCG------AAAC--------- ..(((((.....))))).((((.(((....))).))))....((((......(((..(....)..)))...------.))))...........------....--------- ( -13.80) >DroAna_CAF1 35330 88 + 1 UCGUUAUAAUAAAUAACUUCAACUGUCCG-ACACUUGACACUUGACAUCAACUGCAACGGAAGCGGCA------------GCGGCAAAAAGAA--UGAGAGUC--------- (((((.............((((.((((..-......)))).))))...(..((((..(....)..)))------------)..).......))--))).....--------- ( -17.30) >DroPer_CAF1 44666 99 + 1 UCGUUAUAAUAAAUAACUUCAACUGUCCG-ACCCCUGACACUUGACAUCAACUGCAACGGAAGCGGCAACAGCAACAGGAGAAGCAAAAAAAU---AAGCUUA--------- ..(((((.....))))).((((.((((.(-....).)))).)))).......(((..(....)..))).............((((........---..)))).--------- ( -16.20) >consensus UCGUUAUAAUAAAUAACUUCAACUGUCCA_ACACUUGACACUUGACAUCAACUGCAACGGAAGCGGCA_CA_________G__GCAAAAAGAA_____GAGUC_________ ..(((((.....))))).((((.((((.........)))).)))).......(((..(....)..)))............................................ (-10.48 = -10.65 + 0.17)

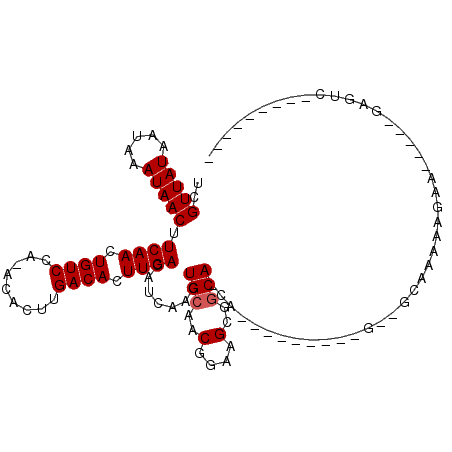
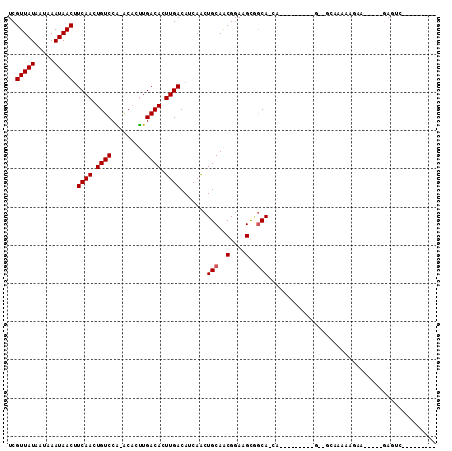
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:25:56 2006