| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 15,199,617 – 15,199,743 |
| Length | 126 |
| Max. P | 0.998971 |

| Location | 15,199,617 – 15,199,719 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.58 |
| Mean single sequence MFE | -30.79 |
| Consensus MFE | -27.38 |
| Energy contribution | -27.38 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -5.07 |
| Structure conservation index | 0.89 |
| SVM decision value | 3.31 |
| SVM RNA-class probability | 0.998971 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
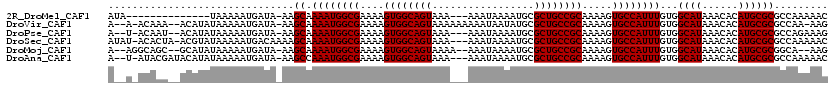
>2R_DroMel_CAF1 15199617 102 + 20766785 AUA--------------UAAAAAUGAUA-AAGCAAAAUGGCGAAAAGUGGCAGUAAA---AAAUAAAAUGCGCUGCCGCAAAAGUGCCAUUUGUGGCAUAAACACAUGCGCGCCAAAAAC ...--------------...........-........(((((....((((((((...---...........))))))))....((((((....))))))...........)))))..... ( -28.94) >DroVir_CAF1 37851 113 + 1 A--A-ACAAA--ACAUAUAAAAAUGAUA-AAGCAAAAUGGCGAAAAGUGGCAGUAAAAAAAAAUAAUAUGCGCUGCCGCAAAAGUGCCAUUUGUGGCAUAAACACAUGCGCGCCAA-AAG .--.-.....--................-........(((((....((((((((.................))))))))....((((((....))))))...........))))).-... ( -28.73) >DroPse_CAF1 35851 111 + 1 A--U-ACAAU--ACAUAUAAAAAUGAUA-AAGCAAAAUGGCGAAAAGUGGCAGUAAA---AAAUAAAAUGCGCUGCCGCAAAAGUGCCAUUUGUGGCAUAAACACAUGCGCGCCAGAAAG .--.-.....--................-........(((((....((((((((...---...........))))))))....((((((....))))))...........)))))..... ( -28.94) >DroSec_CAF1 29748 115 + 1 AUAU-ACACUA-ACGUAUAAAAAUGACAAAAGCAAAAUGGCGAAAAGUGGCAGUAAA---AAAUAAAAUGCGCUGCCGCAAAAGUGCCAUUUGUGGCAUAAACACAUGCGCGCCAAAAAC .(((-((....-..)))))..................(((((....((((((((...---...........))))))))....((((((....))))))...........)))))..... ( -31.54) >DroMoj_CAF1 37458 111 + 1 A--AGGCAGC--GCAUAUAAAAAUGAUA-AAGCAAAAUGGCGAAAAGUGGCAGUAAAA--AAAUAAAAUGCGCUGCCGCAAAAGUGCCAUUUGUGGCAUAAACACAUGCGCGGCA--AAG .--..((.((--((((............-.....((((((((....((((((((....--...........)))))))).....))))))))(((.......))))))))).)).--... ( -37.26) >DroAna_CAF1 25480 113 + 1 A--U-AUACGAUACAUAUAAAAAUGAUA-AAGCCAAAUGGCGAAAAGUGGCAGUAAA---AAAUAAAAUGCGCUGCCGCAAAAGUGCCAUUUGUGGCAUAAACACAUGCGCGCCAAAAAC .--.-.......................-..(((((((((((....((((((((...---...........)))))))).....)))))))))..((((......))))))......... ( -29.34) >consensus A__U_ACAAU__ACAUAUAAAAAUGAUA_AAGCAAAAUGGCGAAAAGUGGCAGUAAA___AAAUAAAAUGCGCUGCCGCAAAAGUGCCAUUUGUGGCAUAAACACAUGCGCGCCAAAAAC ...............................((.((((((((....((((((((.................)))))))).....))))))))...((((......))))))......... (-27.38 = -27.38 + 0.00)
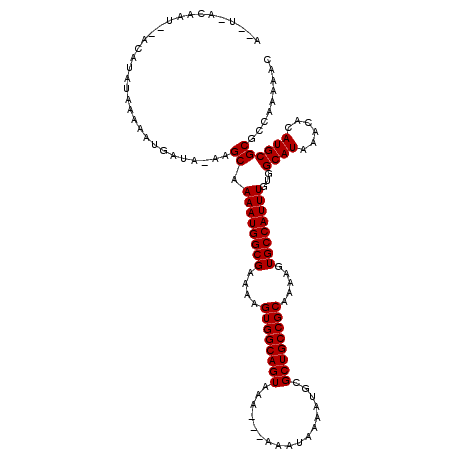

| Location | 15,199,617 – 15,199,719 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.58 |
| Mean single sequence MFE | -25.10 |
| Consensus MFE | -20.71 |
| Energy contribution | -20.71 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.59 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.894552 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15199617 102 - 20766785 GUUUUUGGCGCGCAUGUGUUUAUGCCACAAAUGGCACUUUUGCGGCAGCGCAUUUUAUUU---UUUACUGCCACUUUUCGCCAUUUUGCUU-UAUCAUUUUUA--------------UAU ((...(((((.(((...((....((((....))))))...)))(((((............---....)))))......)))))....))..-...........--------------... ( -22.69) >DroVir_CAF1 37851 113 - 1 CUU-UUGGCGCGCAUGUGUUUAUGCCACAAAUGGCACUUUUGCGGCAGCGCAUAUUAUUUUUUUUUACUGCCACUUUUCGCCAUUUUGCUU-UAUCAUUUUUAUAUGU--UUUGU-U--U ...-.(((((.(((...((....((((....))))))...)))(((((...................)))))......)))))........-................--.....-.--. ( -21.81) >DroPse_CAF1 35851 111 - 1 CUUUCUGGCGCGCAUGUGUUUAUGCCACAAAUGGCACUUUUGCGGCAGCGCAUUUUAUUU---UUUACUGCCACUUUUCGCCAUUUUGCUU-UAUCAUUUUUAUAUGU--AUUGU-A--U .....(((((.(((...((....((((....))))))...)))(((((............---....)))))......)))))...(((..-((.(((......))))--)..))-)--. ( -23.89) >DroSec_CAF1 29748 115 - 1 GUUUUUGGCGCGCAUGUGUUUAUGCCACAAAUGGCACUUUUGCGGCAGCGCAUUUUAUUU---UUUACUGCCACUUUUCGCCAUUUUGCUUUUGUCAUUUUUAUACGU-UAGUGU-AUAU ((...(((((.(((...((....((((....))))))...)))(((((............---....)))))......)))))....))............(((((..-..))))-)... ( -25.49) >DroMoj_CAF1 37458 111 - 1 CUU--UGCCGCGCAUGUGUUUAUGCCACAAAUGGCACUUUUGCGGCAGCGCAUUUUAUUU--UUUUACUGCCACUUUUCGCCAUUUUGCUU-UAUCAUUUUUAUAUGC--GCUGCCU--U ...--.((.(((((((((...(((....(((((((......(.(((((............--.....))))).).....))))))).....-...)))...)))))))--)).))..--. ( -30.03) >DroAna_CAF1 25480 113 - 1 GUUUUUGGCGCGCAUGUGUUUAUGCCACAAAUGGCACUUUUGCGGCAGCGCAUUUUAUUU---UUUACUGCCACUUUUCGCCAUUUGGCUU-UAUCAUUUUUAUAUGUAUCGUAU-A--U ......((((.(((((....)))))).((((((((......(.(((((............---....))))).).....))))))))))).-.........((((((...)))))-)--. ( -26.69) >consensus CUUUUUGGCGCGCAUGUGUUUAUGCCACAAAUGGCACUUUUGCGGCAGCGCAUUUUAUUU___UUUACUGCCACUUUUCGCCAUUUUGCUU_UAUCAUUUUUAUAUGU__AGUGU_A__U .........(((((((....)))))...(((((((......(.(((((...................))))).).....))))))).))............................... (-20.71 = -20.71 + 0.00)

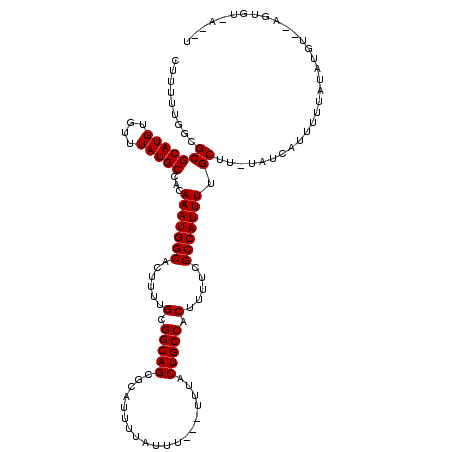
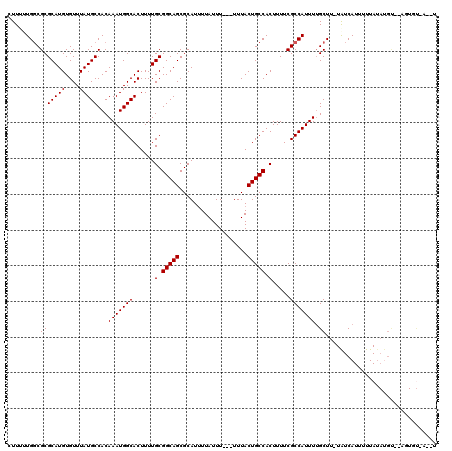
| Location | 15,199,642 – 15,199,743 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.55 |
| Mean single sequence MFE | -26.20 |
| Consensus MFE | -21.81 |
| Energy contribution | -21.81 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.80 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.28 |
| SVM RNA-class probability | 0.939426 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
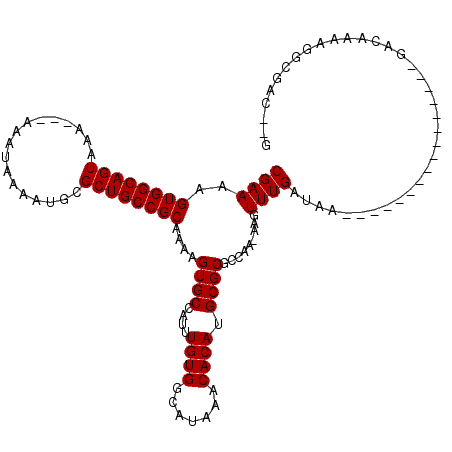
>2R_DroMel_CAF1 15199642 101 + 20766785 CGAAAAGUGGCAGUAAA---AAAUAAAAUGCGCUGCCGCAAAAGUGCCAUUUGUGGCAUAAACACAUGCGCGCCAAAAACUUUGAUAA----------------GACAAAAGCCGAGCCG ......((((((((...---...........))))))))....((((((....)))))).........((.((......(((....))----------------)......))))..... ( -22.54) >DroVir_CAF1 37885 102 + 1 CGAAAAGUGGCAGUAAAAAAAAAUAAUAUGCGCUGCCGCAAAAGUGCCAUUUGUGGCAUAAACACAUGCGCGCCAA-AAGUUUGAUAA----------------GACAAAAGGCGAC-GG ......((((((((.................))))))))....((((((....)))))).........((((((..-..((((....)----------------)))....)))).)-). ( -30.43) >DroGri_CAF1 29093 98 + 1 CGAAAAGUGGCAGUAAAA--AAAUAAAAUGCGCUGCCGCAAAAGUGCCAUUUGUGGCAUAAACACAUGCGCGCCAA-AAGUUUGAUAA----------------GACAAAAGGCGAC--- ......((((((((....--...........))))))))....((((((....))))))...........((((..-..((((....)----------------)))....))))..--- ( -28.36) >DroWil_CAF1 80362 116 + 1 CGAAAAGUGGCAGUAAA---AAAUAAAAUGCGCUGCCGCAAAAGUGCCAUUUGUGGCAUAAACACAUGCGCGCCAAAAAGUUUGAUAACACACAUACACACAAAGACAAAAGACGAU-GG ((....((((((((...---...........))))))))....(((.....((((((((......))))..........(((....))).))))....)))............))..-.. ( -23.44) >DroMoj_CAF1 37493 93 + 1 CGAAAAGUGGCAGUAAAA--AAAUAAAAUGCGCUGCCGCAAAAGUGCCAUUUGUGGCAUAAACACAUGCGCGGCA--AAGUUUGAUAA----------------GACAAAAGG------- ......((((((((....--...........)))))))).....((((...((((.......)))).....))))--..((((....)----------------)))......------- ( -23.26) >DroPer_CAF1 34314 101 + 1 CGAAAAGUGGCAGUAAA---AAAUAAAAUGCGCUGCCGCAAAAGUGCCAUUUGUGGCAUAAACACAUGCGCGCCAGAAAGUUUGAUAA----------------GACAAAGGCAGUGGCA ......((((((((...---...........)))))))).....(((((((.(((.(((......))))))(((.....((((....)----------------)))...)))))))))) ( -29.14) >consensus CGAAAAGUGGCAGUAAA___AAAUAAAAUGCGCUGCCGCAAAAGUGCCAUUUGUGGCAUAAACACAUGCGCGCCAA_AAGUUUGAUAA________________GACAAAAGGCGAC__G ((((..((((((((.................))))))))....((((....((((.......)))).)))).........)))).................................... (-21.81 = -21.81 + -0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:25:55 2006