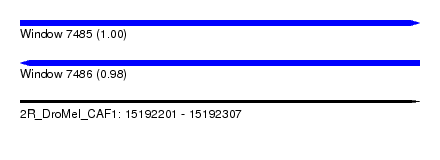
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 15,192,201 – 15,192,307 |
| Length | 106 |
| Max. P | 0.998736 |

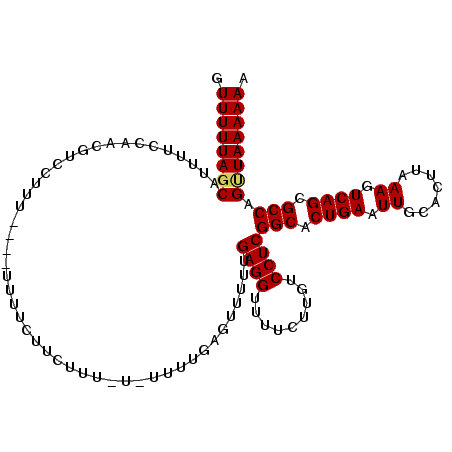
| Location | 15,192,201 – 15,192,307 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 91.92 |
| Mean single sequence MFE | -22.34 |
| Consensus MFE | -21.88 |
| Energy contribution | -21.95 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.24 |
| Structure conservation index | 0.98 |
| SVM decision value | 3.21 |
| SVM RNA-class probability | 0.998736 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15192201 106 + 20766785 UUUUUUAACUGGCGCUGACUUUAAGUGCAAUUCAGUGCCGAGGACAAGAAAACCUCAAAAACUCAAAA-A-AAAGAAGAAAA----AAAGGACGUUGGAAAAUGGUAAAAAC (((((((((.((((((((.............))))))))((((.........))))............-.-...........----.......))))))))).......... ( -22.42) >DroSec_CAF1 22394 108 + 1 UUUUUUAGCUGGCGCUGACUUUAAGUGCAAUUCAGUGCCGAGGACAAGAAAACCUCAAAAACUAAAAAAAAAAAGAAGAAAA----AAAGAACGUUGGAAAAUGCUAAAAAC .((((((((.((((((((.............))))))))((((.........))))..........................----.................)))))))). ( -24.02) >DroSim_CAF1 22468 106 + 1 UUUUUUAACUGGCGCUGACUUUAAGUGCAAUUCAGUGCCGAGGACAAGAAAACCUCAAAAACUCAAAA-AAAAAGAAGAAAA-----AAGGACGUUGGAAAAUGCUAAAAAC (((((((((.((((((((.............))))))))((((.........))))............-.............-----......))))))))).......... ( -22.42) >DroEre_CAF1 22456 109 + 1 UUUUUUAACUGGCGCUGACUUUAAGUGCAAUUCAGUGCCGAGGACAAGAAAACCUCAAAAC-UCAAAA-A-AAAAAAAAAAACAAAAAGGAACGUCGGAAAAUGCUAAAAAC (((((..((.((((((((.............))))))))((((.........)))).....-......-.-......................))..))))).......... ( -20.52) >consensus UUUUUUAACUGGCGCUGACUUUAAGUGCAAUUCAGUGCCGAGGACAAGAAAACCUCAAAAACUCAAAA_A_AAAGAAGAAAA____AAAGAACGUUGGAAAAUGCUAAAAAC (((((((((.((((((((.............))))))))((((.........)))).....................................))))))))).......... (-21.88 = -21.95 + 0.06)

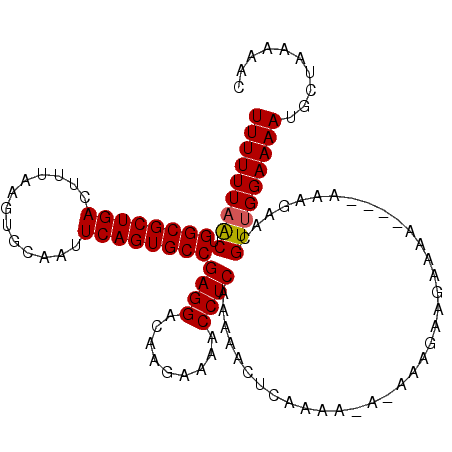
| Location | 15,192,201 – 15,192,307 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 91.92 |
| Mean single sequence MFE | -20.55 |
| Consensus MFE | -20.50 |
| Energy contribution | -20.62 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.99 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.91 |
| SVM RNA-class probability | 0.982383 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15192201 106 - 20766785 GUUUUUACCAUUUUCCAACGUCCUUU----UUUUCUUCUUU-U-UUUUGAGUUUUUGAGGUUUUCUUGUCCUCGGCACUGAAUUGCACUUAAAGUCAGCGCCAGUUAAAAAA ..........................----..........(-(-((((((......((((.........))))(((.((((.((.......)).)))).)))..)))))))) ( -17.40) >DroSec_CAF1 22394 108 - 1 GUUUUUAGCAUUUUCCAACGUUCUUU----UUUUCUUCUUUUUUUUUUUAGUUUUUGAGGUUUUCUUGUCCUCGGCACUGAAUUGCACUUAAAGUCAGCGCCAGCUAAAAAA .((((((((.................----..........................((((.........))))(((.((((.((.......)).)))).))).)))))))). ( -23.20) >DroSim_CAF1 22468 106 - 1 GUUUUUAGCAUUUUCCAACGUCCUU-----UUUUCUUCUUUUU-UUUUGAGUUUUUGAGGUUUUCUUGUCCUCGGCACUGAAUUGCACUUAAAGUCAGCGCCAGUUAAAAAA .((((((((................-----.............-............((((.........))))(((.((((.((.......)).)))).))).)))))))). ( -20.80) >DroEre_CAF1 22456 109 - 1 GUUUUUAGCAUUUUCCGACGUUCCUUUUUGUUUUUUUUUUU-U-UUUUGA-GUUUUGAGGUUUUCUUGUCCUCGGCACUGAAUUGCACUUAAAGUCAGCGCCAGUUAAAAAA .((((((((................................-.-......-.....((((.........))))(((.((((.((.......)).)))).))).)))))))). ( -20.80) >consensus GUUUUUAGCAUUUUCCAACGUCCUUU____UUUUCUUCUUU_U_UUUUGAGUUUUUGAGGUUUUCUUGUCCUCGGCACUGAAUUGCACUUAAAGUCAGCGCCAGUUAAAAAA .((((((((...............................................((((.........))))(((.((((.((.......)).)))).))).)))))))). (-20.50 = -20.62 + 0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:25:52 2006