| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 15,185,052 – 15,185,148 |
| Length | 96 |
| Max. P | 0.839173 |

| Location | 15,185,052 – 15,185,148 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 82.49 |
| Mean single sequence MFE | -23.47 |
| Consensus MFE | -13.94 |
| Energy contribution | -15.75 |
| Covariance contribution | 1.81 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.791647 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
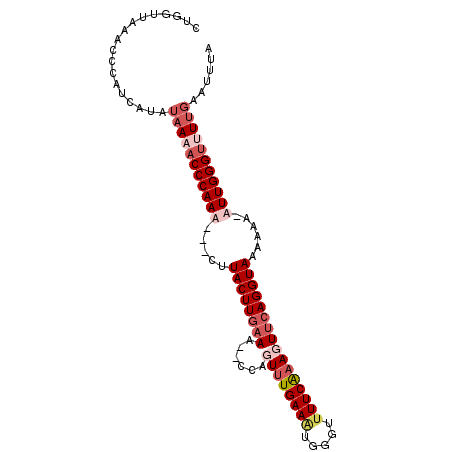
>2R_DroMel_CAF1 15185052 96 + 20766785 UAAAUUCAAAACCCAAU-UUUCUUACCUGAAAUUUGAAAACCCAUUUCAAACUGGGAUUUCAAGUAUG---UUUGGGUUUUAUAUGACGGGUCUAACCAG .......(((((((((.-.(((......)))((((((((.((((........)))).))))))))...---.))))))))).......((......)).. ( -25.40) >DroSim_CAF1 15468 95 + 1 UAAAUUCAAAACCCAAU-UUUCUUACCUGUAAU-UGAAAACCCAUUUCAAACGGGGAUUUUAAGUAAG---UUUGGGUUUUAUAUGACGGGUCUUACCAG .......(((((((((.-((((((((((((..(-(((((.....))))))))))).....)))).)))---.))))))))).......((......)).. ( -22.60) >DroEre_CAF1 15528 98 + 1 UAAAUUCAAGGCCCAAAGUUUUAUACCUGAACUUUGAAAACCCACUUCAAACUGG--UUUCAAGUAAGUGUUUUGGGUUUUAUGUGUUGGGCUUAACCAG ........(((((((((((((.......))))))).((((((((...((.(((..--.....)))...))...)))))))).......))))))...... ( -25.10) >DroYak_CAF1 15518 98 + 1 UAAAUUGCAUACCCAAAGUUUUGUACCUGAACUUCGAAAACCCAGUUCAAACUGG--UUUCAAGUAAGUAGUUUGGGUUUUAUAUGAUGGGCUUAAACAG ...........((((..((.....(((..((((.(((((..(((((....)))))--))))......).))))..))).....))..))))......... ( -20.80) >consensus UAAAUUCAAAACCCAAA_UUUCUUACCUGAAAUUUGAAAACCCAUUUCAAACUGG__UUUCAAGUAAG___UUUGGGUUUUAUAUGACGGGCCUAACCAG ..........((((..........(((((((((((((((.((((........)))).))))))))......)))))))..........))))........ (-13.94 = -15.75 + 1.81)


| Location | 15,185,052 – 15,185,148 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 82.49 |
| Mean single sequence MFE | -21.50 |
| Consensus MFE | -15.04 |
| Energy contribution | -17.98 |
| Covariance contribution | 2.94 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.839173 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15185052 96 - 20766785 CUGGUUAGACCCGUCAUAUAAAACCCAAA---CAUACUUGAAAUCCCAGUUUGAAAUGGGUUUUCAAAUUUCAGGUAAGAAA-AUUGGGUUUUGAAUUUA ..((......))......((((((((((.---.....((((((.((((........)))).)))))).((((......))))-.))))))))))...... ( -24.40) >DroSim_CAF1 15468 95 - 1 CUGGUAAGACCCGUCAUAUAAAACCCAAA---CUUACUUAAAAUCCCCGUUUGAAAUGGGUUUUCA-AUUACAGGUAAGAAA-AUUGGGUUUUGAAUUUA ..((......))......((((((((((.---(((((((..((..(((((.....)))))..))..-.....)))))))...-.))))))))))...... ( -20.80) >DroEre_CAF1 15528 98 - 1 CUGGUUAAGCCCAACACAUAAAACCCAAAACACUUACUUGAAA--CCAGUUUGAAGUGGGUUUUCAAAGUUCAGGUAUAAAACUUUGGGCCUUGAAUUUA ....((((((((((........(((...(((((((((((.(((--....))).)))))))).......)))..)))........))))).)))))..... ( -20.20) >DroYak_CAF1 15518 98 - 1 CUGUUUAAGCCCAUCAUAUAAAACCCAAACUACUUACUUGAAA--CCAGUUUGAACUGGGUUUUCGAAGUUCAGGUACAAAACUUUGGGUAUGCAAUUUA ......................(((((((.....((((((((.--(((((....)))))..........))))))))......))))))).......... ( -20.60) >consensus CUGGUUAAACCCAUCAUAUAAAACCCAAA___CUUACUUGAAA__CCAGUUUGAAAUGGGUUUUCAAAGUUCAGGUAAAAAA_AUUGGGUUUUGAAUUUA ..................(((((((((((.....((((((((......((((((((.....))))))))))))))))......)))))))))))...... (-15.04 = -17.98 + 2.94)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:25:50 2006