| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 15,181,216 – 15,181,324 |
| Length | 108 |
| Max. P | 0.804829 |

| Location | 15,181,216 – 15,181,324 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 78.12 |
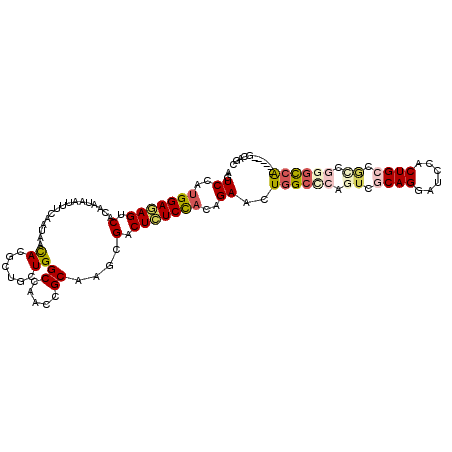
| Mean single sequence MFE | -34.53 |
| Consensus MFE | -19.25 |
| Energy contribution | -20.08 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.804829 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
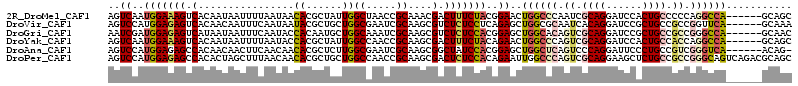
>2R_DroMel_CAF1 15181216 108 - 20766785 AGUCAAUGGAAAGUCACAAUAAUUUUAAUAACACGCUAUUGGCUAACCGCAAACGACUUUCUACGGAACUGGCCCAAUCGCAGGAUCCACUGCCCCCAGGCCA------GCAGC ..((..(((((((((.(((((...............)))))((.....))....)))))))))..)).((((((.....((((......)))).....)))))------).... ( -32.36) >DroVir_CAF1 9898 108 - 1 AGUCCAUGGAGAGUCACAACAAUUUCAAUAAUACGCUGCUGGCGAAUCGCAAGCGUCUCUCCUCAGAGCUGGCGCAAUCACAGGAUCCGCUGCCGCCGGUUCA------GCAAA .......((((((...................(((((....(((...))).)))))))))))...((((((((((((((....)))....)).))))))))).------..... ( -32.20) >DroGri_CAF1 8028 108 - 1 AAUCGAUGGAGAGUCAUAAUAAUUUCAAUACCACAAUGCUGGCAAAUCGCAAGCGUCUCUCCACGGAGCUGGCACAGUCGCAGGAUCCGCUGCCGCCGGGCCA------GCAAC ..(((.(((((((......................(((((.((.....)).))))))))))))))).((((((.(.((.((((......)))).)).).))))------))... ( -37.94) >DroYak_CAF1 11704 108 - 1 AGUCAAUGGAAAGUCACAAUAAUUUUAAUACCACGCUAUUGGCCAACCGCAAGCGACUUUCUACAGAACUGGCCCAGUCGCAGGAUCCACUGCCACCAGGCCA------GCAGC ..((..(((((((((..((.....))........(((..(((....)))..))))))))))))..)).((((((..((.((((......)))).))..)))))------).... ( -33.70) >DroAna_CAF1 8649 107 - 1 AGUCCAUGGAGAGCCACAACAACUUCAACAACACGCUCUUGGCGAAUCGCAAGCGGCUAUCCACGGAGCUGGCUCAGUCCCAGGAUUCCCUGCCGUCGGGUCA------ACAG- ..(((.((((.((((..................(((.....)))...(....).)))).)))).)))(.((((((..(..((((....))))..)..))))))------.)..- ( -33.10) >DroPer_CAF1 15926 114 - 1 AGUCCAUGGAGAGCCACACUAGCUUUAACAACACGCUGCUGGCCAACCGCAAGCGACUCUCCACAGAAUUGGCCCAGUCGCAGGAAGCUCUGCCGCCGGGCAGUCAGACGCAGC .(((..(((((((.(.....(((...........)))(((.((.....)).)))).)))))))..((....((((.((.(((((....))))).)).))))..)).)))..... ( -37.90) >consensus AGUCCAUGGAGAGUCACAAUAAUUUCAAUAACACGCUGCUGGCCAACCGCAAGCGACUCUCCACAGAACUGGCCCAGUCGCAGGAUCCACUGCCGCCGGGCCA______GCAGC ..((..(((((((.(................((......))((.....))....).)))))))..))..((((((.((.((((......)))).)).))))))........... (-19.25 = -20.08 + 0.84)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:25:47 2006