
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 15,174,938 – 15,175,091 |
| Length | 153 |
| Max. P | 0.981987 |

| Location | 15,174,938 – 15,175,028 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 82.84 |
| Mean single sequence MFE | -34.52 |
| Consensus MFE | -19.89 |
| Energy contribution | -19.75 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.556608 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
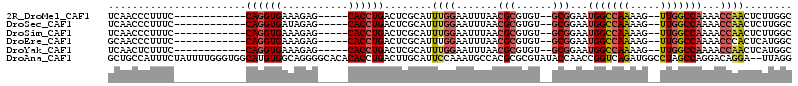
>2R_DroMel_CAF1 15174938 90 + 20766785 GCAAA--CUGGC---CACG-CCU-CGGGCCAAGAGUUGGUUUUGGCCAA--CUUUUGGCCAUUCCGC--ACACGCGUUAAAUUCCAAAUGCGAGUCAGGUG- .....--(((((---...(-(..-..((((((((((((((....)))))--))))))))).....))--...((((((........)))))).)))))...- ( -39.00) >DroSec_CAF1 5330 90 + 1 GCAAA--CUGGC---CACG-CCU-CGGGCCAAGAGUUGGUUUUGGCCAA--CUUUUGGCCAUUCCGC--ACACGCGUUAAAUUCCAAAUGCGAGUCAGGUG- .....--(((((---...(-(..-..((((((((((((((....)))))--))))))))).....))--...((((((........)))))).)))))...- ( -39.00) >DroSim_CAF1 5372 90 + 1 GCAAA--CUGGC---CACG-CCU-CGGGCCAAGAGUUGGUUUUGGCCAA--CUUUUGGCCAUUCCGC--ACACGCGUUAAAUUCCAAAUGCGAGUCAGGUG- .....--(((((---...(-(..-..((((((((((((((....)))))--))))))))).....))--...((((((........)))))).)))))...- ( -39.00) >DroEre_CAF1 5391 93 + 1 GCAAA--CUGGCCGCCACG-CCU-CGGGCCAUGAGUGGGUUUUGGCCAA--CUUUUGGCCAUUCCGC--ACACGCGUUAAAUUCCAAAUGCGAGUCAGGUG- .....--(((((.(((.((-...-)))))..((.(((((...((((((.--....)))))).)))))--.))((((((........)))))).)))))...- ( -32.90) >DroYak_CAF1 5350 91 + 1 GCAAA--CUGGC---CACG-CCUGUGGGCCAUGAGUUGGUUUUGGCCAA--CUUUUGGCCAUUCCGC--ACACGCGUUAAAUUCCAAAUGCGAGUCAGGUG- .....--(((((---...(-(.....(((((.((((((((....)))))--))).))))).....))--...((((((........)))))).)))))...- ( -36.20) >DroAna_CAF1 1486 96 + 1 GCAAAAGCAGUC---CACAACCU-CAUCCUAA--UCCUGUCCUGGCUAGGCCAUCUGACCGGUUGGUAUACGCGCGUGGCAUUUGGAAUGCAAGUCAGGUGU ......(((.((---((......-....((((--.((.(((.((((...))))...))).)))))).....((.....))...)))).)))........... ( -21.00) >consensus GCAAA__CUGGC___CACG_CCU_CGGGCCAAGAGUUGGUUUUGGCCAA__CUUUUGGCCAUUCCGC__ACACGCGUUAAAUUCCAAAUGCGAGUCAGGUG_ .......(((((.........................((...(((((((.....)))))))..)).......((((((........)))))).))))).... (-19.89 = -19.75 + -0.14)

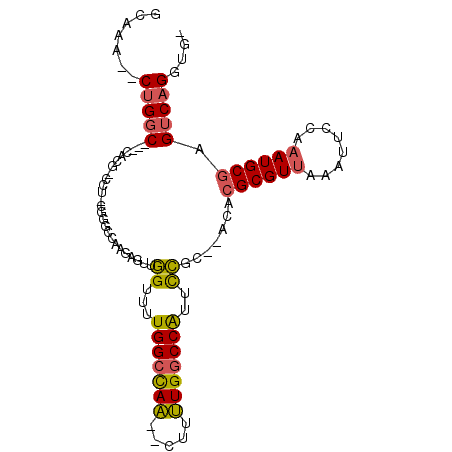
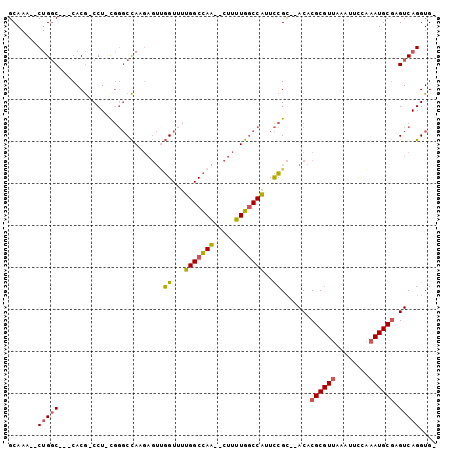
| Location | 15,174,958 – 15,175,051 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 79.40 |
| Mean single sequence MFE | -36.84 |
| Consensus MFE | -21.50 |
| Energy contribution | -21.45 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.30 |
| Mean z-score | -3.27 |
| Structure conservation index | 0.58 |
| SVM decision value | 1.90 |
| SVM RNA-class probability | 0.981987 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
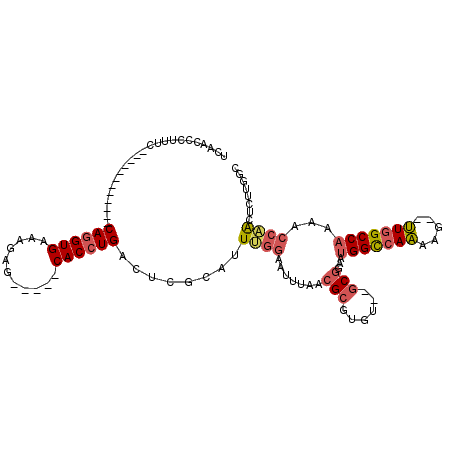
>2R_DroMel_CAF1 15174958 93 + 20766785 GCCAAGAGUUGGUUUUGGCCAA--CUUUUGGCCAUUCCGC--ACACGCGUUAAAUUCCAAAUGCGAGUCAGGUG-----CUCUUUCACCUG------------GAAAGGGUUGA (((((((((((((....)))))--))))))))((..((..--...((((((........))))))..(((((((-----......))))))------------)...))..)). ( -39.40) >DroSec_CAF1 5350 93 + 1 GCCAAGAGUUGGUUUUGGCCAA--CUUUUGGCCAUUCCGC--ACACGCGUUAAAUUCCAAAUGCGAGUCAGGUG-----CUCUAUCACCUG------------GAAAGGGUUGA (((((((((((((....)))))--))))))))((..((..--...((((((........))))))..(((((((-----......))))))------------)...))..)). ( -39.40) >DroSim_CAF1 5392 93 + 1 GCCAAGAGUUGGUUUUGGCCAA--CUUUUGGCCAUUCCGC--ACACGCGUUAAAUUCCAAAUGCGAGUCAGGUG-----CUCUUUCACCUG------------GAAAGGGUUGA (((((((((((((....)))))--))))))))((..((..--...((((((........))))))..(((((((-----......))))))------------)...))..)). ( -39.40) >DroEre_CAF1 5414 93 + 1 GCCAUGAGUGGGUUUUGGCCAA--CUUUUGGCCAUUCCGC--ACACGCGUUAAAUUCCAAAUGCGAGUCAGGUG-----CUCUUUCACCUG------------GAAAGGGUUGC (((.((((((((...((((((.--....)))))).)))))--...((((((........))))))..))))))(-----(((((((.....------------))))))))... ( -35.50) >DroYak_CAF1 5371 93 + 1 GCCAUGAGUUGGUUUUGGCCAA--CUUUUGGCCAUUCCGC--ACACGCGUUAAAUUCCAAAUGCGAGUCAGGUG-----CUCUUUCACCUG------------GAAAGAGUUGA ((((.((((((((....)))))--))).))))....((..--((.((((((........)))))).))..)).(-----(((((((.....------------))))))))... ( -34.40) >DroAna_CAF1 1509 112 + 1 CCUAA--UCCUGUCCUGGCUAGGCCAUCUGACCGGUUGGUAUACGCGCGUGGCAUUUGGAAUGCAAGUCAGGUGUGUGCCCCUGCCACAUGCCACCCAAAAUAGAAAUGGCAGC .....--....(((.((((...))))...))).(((.((..(((((((.((((..(((.....))))))).)))))))..)).)))...(((((.............))))).. ( -32.92) >consensus GCCAAGAGUUGGUUUUGGCCAA__CUUUUGGCCAUUCCGC__ACACGCGUUAAAUUCCAAAUGCGAGUCAGGUG_____CUCUUUCACCUG____________GAAAGGGUUGA (((.......((...(((((((.....)))))))..))....((.((((((........)))))).))..)))......(((((((.................))))))).... (-21.50 = -21.45 + -0.05)

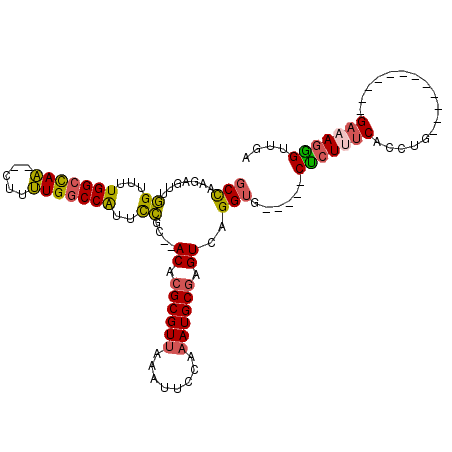
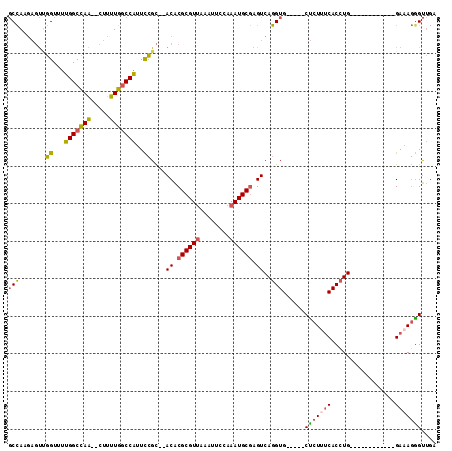
| Location | 15,174,958 – 15,175,051 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 79.40 |
| Mean single sequence MFE | -31.05 |
| Consensus MFE | -18.33 |
| Energy contribution | -18.88 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.788192 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15174958 93 - 20766785 UCAACCCUUUC------------CAGGUGAAAGAG-----CACCUGACUCGCAUUUGGAAUUUAACGCGUGU--GCGGAAUGGCCAAAAG--UUGGCCAAAACCAACUCUUGGC ........(((------------((((((...(((-----(....).))).))))))))).....(((....--))).....(((((.((--((((......)))))).))))) ( -30.80) >DroSec_CAF1 5350 93 - 1 UCAACCCUUUC------------CAGGUGAUAGAG-----CACCUGACUCGCAUUUGGAAUUUAACGCGUGU--GCGGAAUGGCCAAAAG--UUGGCCAAAACCAACUCUUGGC ........(((------------((((((...(((-----(....).))).))))))))).....(((....--))).....(((((.((--((((......)))))).))))) ( -30.80) >DroSim_CAF1 5392 93 - 1 UCAACCCUUUC------------CAGGUGAAAGAG-----CACCUGACUCGCAUUUGGAAUUUAACGCGUGU--GCGGAAUGGCCAAAAG--UUGGCCAAAACCAACUCUUGGC ........(((------------((((((...(((-----(....).))).))))))))).....(((....--))).....(((((.((--((((......)))))).))))) ( -30.80) >DroEre_CAF1 5414 93 - 1 GCAACCCUUUC------------CAGGUGAAAGAG-----CACCUGACUCGCAUUUGGAAUUUAACGCGUGU--GCGGAAUGGCCAAAAG--UUGGCCAAAACCCACUCAUGGC ((......(((------------((((((...(((-----(....).))).)))))))))......))(.((--(.((..((((((....--.))))))...))))).)..... ( -30.80) >DroYak_CAF1 5371 93 - 1 UCAACUCUUUC------------CAGGUGAAAGAG-----CACCUGACUCGCAUUUGGAAUUUAACGCGUGU--GCGGAAUGGCCAAAAG--UUGGCCAAAACCAACUCAUGGC ........(((------------((((((...(((-----(....).))).))))))))).....(((....--))).....((((..((--((((......))))))..)))) ( -29.30) >DroAna_CAF1 1509 112 - 1 GCUGCCAUUUCUAUUUUGGGUGGCAUGUGGCAGGGGCACACACCUGACUUGCAUUCCAAAUGCCACGCGCGUAUACCAACCGGUCAGAUGGCCUAGCCAGGACAGGA--UUAGG ....((..((((.(((((((((((((.(((((((........)))).........))).)))))))((.(((..(((....)))...)))))....)))))).))))--...)) ( -33.80) >consensus UCAACCCUUUC____________CAGGUGAAAGAG_____CACCUGACUCGCAUUUGGAAUUUAACGCGUGU__GCGGAAUGGCCAAAAG__UUGGCCAAAACCAACUCUUGGC .......................((((((...........))))))........((((.......(((......)))...(((((((.....)))))))...))))........ (-18.33 = -18.88 + 0.56)
| Location | 15,174,991 – 15,175,091 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 98.40 |
| Mean single sequence MFE | -31.20 |
| Consensus MFE | -29.80 |
| Energy contribution | -29.84 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.16 |
| Structure conservation index | 0.96 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
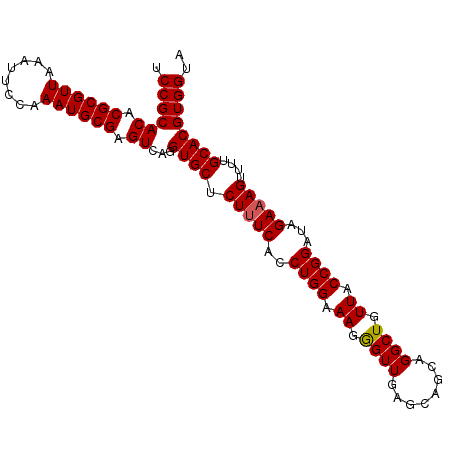
>2R_DroMel_CAF1 15174991 100 + 20766785 UCCGCACACGCGUUAAAUUCCAAAUGCGAGUCAGGUGCUCUUUCACCUGGAAAGGGUUGAGCAGCAGGCUGUUACCGGAUAGAAAGUUUUGCACGUGGUA .((((((.((((((........)))))).))...((((.(((((.(((....)))....(((((....)))))........)))))....)))))))).. ( -31.70) >DroSec_CAF1 5383 100 + 1 UCCGCACACGCGUUAAAUUCCAAAUGCGAGUCAGGUGCUCUAUCACCUGGAAAGGGUUGAGCAGCAGGCUGUUACCGGAUAGAAAGUUUUGCACGUGGUA .((((((.((((((........)))))).))...((((((((((.(((....)))....(((((....)))))....)))))).......)))))))).. ( -31.31) >DroSim_CAF1 5425 100 + 1 UCCGCACACGCGUUAAAUUCCAAAUGCGAGUCAGGUGCUCUUUCACCUGGAAAGGGUUGAGCAGCAGGCUGUUACCGGAUAGAAAGUUUUGCACGUGGUA .((((((.((((((........)))))).))...((((.(((((.(((....)))....(((((....)))))........)))))....)))))))).. ( -31.70) >DroEre_CAF1 5447 100 + 1 UCCGCACACGCGUUAAAUUCCAAAUGCGAGUCAGGUGCUCUUUCACCUGGAAAGGGUUGCCCAGCAGGCUGUUACCGGAUAGAAAGUUUUGCACGUGGUA .((((((.((((((........)))))).))...((((.(((((..((((.((.((((((...))).))).)).))))...)))))....)))))))).. ( -31.10) >DroYak_CAF1 5404 100 + 1 UCCGCACACGCGUUAAAUUCCAAAUGCGAGUCAGGUGCUCUUUCACCUGGAAAGAGUUGAGCAGCAGGCUGUUACCGGAUAGAAAGUUUUGCACGUGGUA .((((((.((((((........)))))).))...((((.(((((..((((.((.((((........)))).)).))))...)))))....)))))))).. ( -30.20) >consensus UCCGCACACGCGUUAAAUUCCAAAUGCGAGUCAGGUGCUCUUUCACCUGGAAAGGGUUGAGCAGCAGGCUGUUACCGGAUAGAAAGUUUUGCACGUGGUA .((((((.((((((........)))))).))...((((.(((((..((((.((.((((........)))).)).))))...)))))....)))))))).. (-29.80 = -29.84 + 0.04)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:25:34 2006