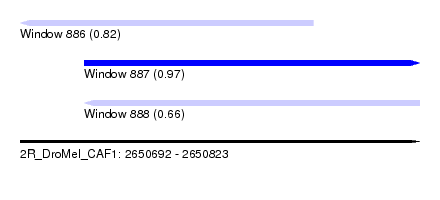
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 2,650,692 – 2,650,823 |
| Length | 131 |
| Max. P | 0.973384 |

| Location | 2,650,692 – 2,650,788 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.64 |
| Mean single sequence MFE | -28.25 |
| Consensus MFE | -16.47 |
| Energy contribution | -15.97 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.820684 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
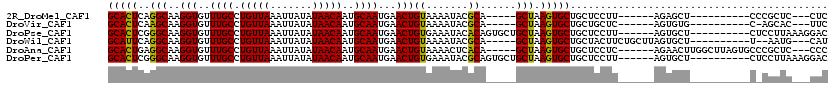
>2R_DroMel_CAF1 2650692 96 - 20766785 GCACUCAGGCAAGGUGUUUGCCUGUUAAAUUAUAUAACAAUGCAAUGAACUGUAAAAUACGCA-----GCUAAGUGCUGCUCCUU------AGAGCU----------CCCGCUC---CUC .....((((((((...))))))))................((((......))))......(((-----((.....))))).....------.((((.----------...))))---... ( -23.90) >DroVir_CAF1 105583 95 - 1 GCACUCAAGCAAGGUGUUUGCCUGUUAAAUUAUAUAACAAUGCAAUGAACUGUAAAAUACGCA-----GCUAAGUGCUGCUGCUC------AGUGUG----------C-AGCAC---UUC ((((.(.((((.(((..((((.(((((.......)))))..))))...))).........(((-----((.....))))))))).------.).)))----------)-.....---... ( -26.30) >DroPse_CAF1 95382 104 - 1 GCACUCGGGCAAGGUGUUUGCCUGUUAAAUUAUAUAACAAUGCAAUGAACUGUGAAAUACACAGUGCUGCUAAGUGCUGCUCCUU------AGUGCU----------CUCCUUAAAGGAC (((((.(((...((..((((..(((((.......)))))..(((....((((((.....))))))..)))))))..))...))).------))))).----------.(((.....))). ( -29.50) >DroWil_CAF1 116315 100 - 1 GCAUUCAGGCAAGGUGUUUGCCUGUUAAAUUAUAUAACAAUGCAAUGAACUGUAAAAUACGCA-----GCUAAGUGCUGCUACUUCUGCUUAGUGCU----------U--AAUG---CAU (((((.(((((.(((..((((.(((((.......)))))..))))...))).........(((-----((.....))))).............))))----------)--))))---).. ( -26.90) >DroAna_CAF1 88327 106 - 1 GCACUGAGGCAAGGUGUUUGCCUGUUAAAUUAUAUAACAAUGCAAUGAACUGUAAAACUCACA-----GCUAAGUGCUGCUCCUC------AGAACUUGGCUUAGUGCCCGCUC---CCC ((((((((.((((((..((((.(((((.......)))))..))))...))...........((-----((.....))))......------....)))).))))))))......---... ( -30.10) >DroPer_CAF1 101624 104 - 1 GCACUCGGGCAAGGUGUUUGCCUGUUAAAUUAUAUAACAAUGCAAUGAACUGUGAAAUACGCAGUGCUGCUAAGUGCUGCUCCUU------AGUGCU----------CUCCUUAAAGGAC (((((.(((((.(((..((((.(((((.......)))))..))))...))).))......((((..((....))..)))).))).------))))).----------.(((.....))). ( -32.80) >consensus GCACUCAGGCAAGGUGUUUGCCUGUUAAAUUAUAUAACAAUGCAAUGAACUGUAAAAUACGCA_____GCUAAGUGCUGCUCCUU______AGUGCU__________C_CGCUC___CAC (((((..(((..(((..((((.(((((.......)))))..))))...)))((.......))......))).)))))........................................... (-16.47 = -15.97 + -0.50)
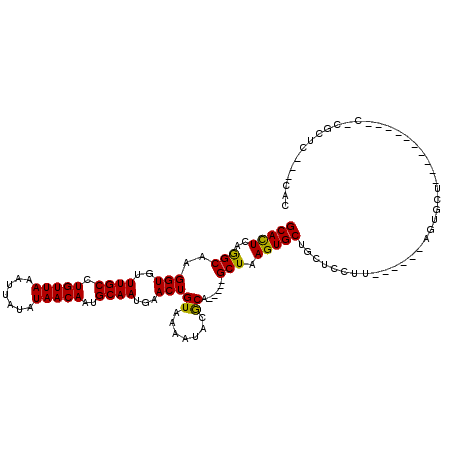
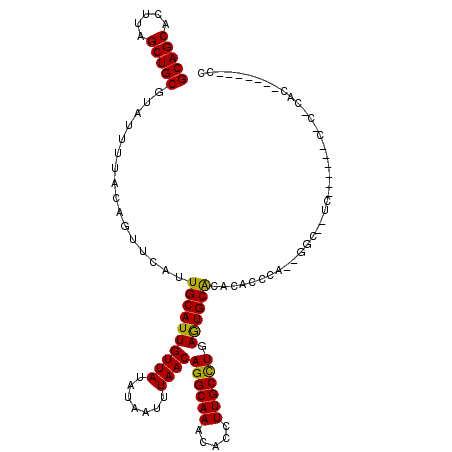
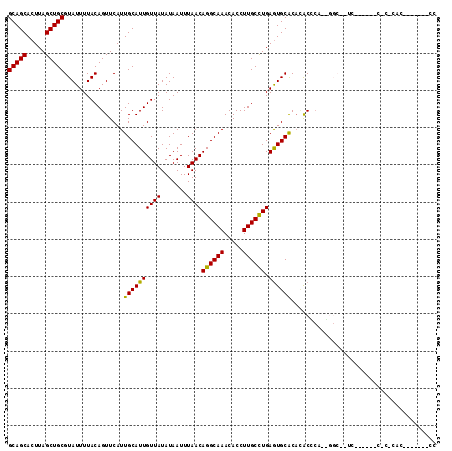
| Location | 2,650,713 – 2,650,823 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 78.42 |
| Mean single sequence MFE | -25.88 |
| Consensus MFE | -21.84 |
| Energy contribution | -21.20 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.64 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.71 |
| SVM RNA-class probability | 0.973384 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2650713 110 + 20766785 GCAGCACUUAGCUGCGUAUUUUACAGUUCAUUGCAUUGUUAUAUAAUUUAACAGGCAAACACCUUGCCUGAGUGCACACACUCAAAGGCAUUCG-----CACACACUCGCAUUCC (((((.....)))))...............((((.((((((.......))))))))))......((((((((((....)))))..)))))...(-----(........))..... ( -29.70) >DroVir_CAF1 105603 90 + 1 GCAGCACUUAGCUGCGUAUUUUACAGUUCAUUGCAUUGUUAUAUAAUUUAACAGGCAAACACCUUGCUUGAGUGCGCCUGC------------------CGCCCAC-------CC (((((.....)))))((((...(((((.......))))).)))).......(((((((.....))))))).(((.((....------------------.)).)))-------.. ( -21.90) >DroGri_CAF1 101390 83 + 1 GCAGCACUUAGCUGCGUAUUUUACAGUUCAUUGCAUUGUUAUAUAAUUUAACAGGCAAACACCUUGCUUGAGUGCGCACGCCC-------------------------------- ((.(((((((((...(((...)))......((((.((((((.......)))))))))).......)).)))))))))......-------------------------------- ( -21.00) >DroEre_CAF1 83835 100 + 1 GCAGCACUUAGCUGCGUAUUUUACAGUUCAUUGCAUUGUUAUAUAAUUUAACAGGCAAACACCUUGCCUGAGUGCACACACUCAAAGGCAUUC---------------GCAUUCC (((((.....)))))((.............((((.((((((.......))))))))))......((((((((((....)))))..)))))...---------------))..... ( -28.80) >DroWil_CAF1 116340 108 + 1 GCAGCACUUAGCUGCGUAUUUUACAGUUCAUUGCAUUGUUAUAUAAUUUAACAGGCAAACACCUUGCCUGAAUGCACACACACACUGGC--UCA-----CACACACACACAUACG (((((.....)))))((......((((....((((((((((.......))))((((((.....)))))).)))))).......))))..--...-----.))............. ( -25.50) >DroMoj_CAF1 106924 107 + 1 GCAGCACUUAGCUGCGUAUUUUACAGUUCAUUGCAUUGUUAUAUAAUUUAACAGGCAAACACCUUGCUUGAGUGCGCAAACCCC-GGGCACCGACAUCUCGCCAAG-------CC (((((.....)))))...............((((.((((((.......)))))))))).......(((((.((((.(.......-).))))(((....))).))))-------). ( -28.40) >consensus GCAGCACUUAGCUGCGUAUUUUACAGUUCAUUGCAUUGUUAUAUAAUUUAACAGGCAAACACCUUGCCUGAGUGCACACACCCA__GGC__UC______C_C_CAC_______CC (((((.....)))))................((((((((((.......))))((((((.....)))))).))))))....................................... (-21.84 = -21.20 + -0.64)

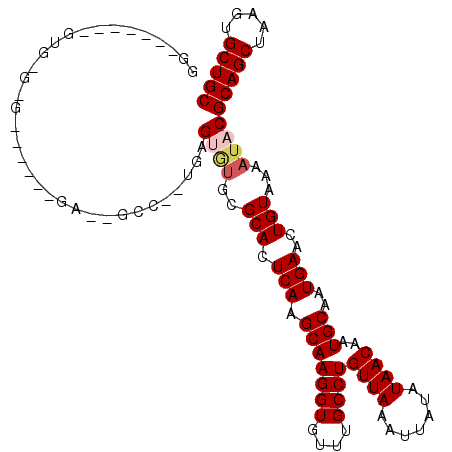
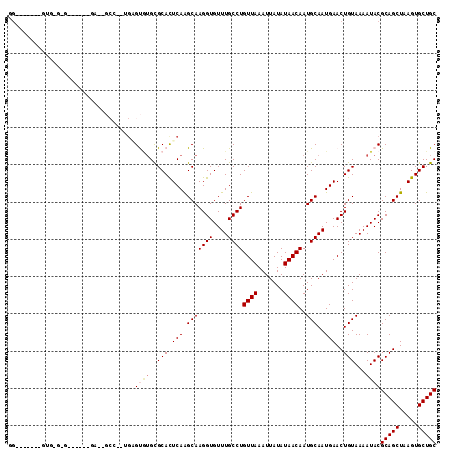
| Location | 2,650,713 – 2,650,823 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 78.42 |
| Mean single sequence MFE | -28.87 |
| Consensus MFE | -16.93 |
| Energy contribution | -17.48 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.658650 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2650713 110 - 20766785 GGAAUGCGAGUGUGUG-----CGAAUGCCUUUGAGUGUGUGCACUCAGGCAAGGUGUUUGCCUGUUAAAUUAUAUAACAAUGCAAUGAACUGUAAAAUACGCAGCUAAGUGCUGC ....(((.(((.((((-----((((((((((((((((....))))))...))))))))))).(((((.......)))))..)))....))))))......(((((.....))))) ( -34.60) >DroVir_CAF1 105603 90 - 1 GG-------GUGGGCG------------------GCAGGCGCACUCAAGCAAGGUGUUUGCCUGUUAAAUUAUAUAACAAUGCAAUGAACUGUAAAAUACGCAGCUAAGUGCUGC .(-------((..(((------------------((((((((.((......)))))))))))(((((.......)))))..)).....))).........(((((.....))))) ( -26.40) >DroGri_CAF1 101390 83 - 1 --------------------------------GGGCGUGCGCACUCAAGCAAGGUGUUUGCCUGUUAAAUUAUAUAACAAUGCAAUGAACUGUAAAAUACGCAGCUAAGUGCUGC --------------------------------(((((.((((.((......)))))).)))))((((.......))))..((((......))))......(((((.....))))) ( -21.00) >DroEre_CAF1 83835 100 - 1 GGAAUGC---------------GAAUGCCUUUGAGUGUGUGCACUCAGGCAAGGUGUUUGCCUGUUAAAUUAUAUAACAAUGCAAUGAACUGUAAAAUACGCAGCUAAGUGCUGC .....((---------------(((((((((((((((....))))))...))))))))))).(((((.......))))).((((......))))......(((((.....))))) ( -31.30) >DroWil_CAF1 116340 108 - 1 CGUAUGUGUGUGUGUG-----UGA--GCCAGUGUGUGUGUGCAUUCAGGCAAGGUGUUUGCCUGUUAAAUUAUAUAACAAUGCAAUGAACUGUAAAAUACGCAGCUAAGUGCUGC .((((..((.((((((-----(..--..((((.......((((((((((((((...))))))))..............))))))....))))....)))))))))...))))... ( -28.43) >DroMoj_CAF1 106924 107 - 1 GG-------CUUGGCGAGAUGUCGGUGCCC-GGGGUUUGCGCACUCAAGCAAGGUGUUUGCCUGUUAAAUUAUAUAACAAUGCAAUGAACUGUAAAAUACGCAGCUAAGUGCUGC .(-------(((((((((((.((((...))-)).)))).)))...)))))..(((..((((.(((((.......)))))..))))...))).........(((((.....))))) ( -31.50) >consensus GG_______GUG_G_G______GA__GCC__UGAGUGUGCGCACUCAAGCAAGGUGUUUGCCUGUUAAAUUAUAUAACAAUGCAAUGAACUGUAAAAUACGCAGCUAAGUGCUGC ..................................((((..(((.(((.(((((((....))))((((.......))))..)))..)))..)))...))))(((((.....))))) (-16.93 = -17.48 + 0.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:39:17 2006