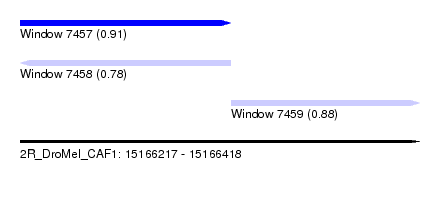
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 15,166,217 – 15,166,418 |
| Length | 201 |
| Max. P | 0.913759 |

| Location | 15,166,217 – 15,166,323 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 93.39 |
| Mean single sequence MFE | -29.26 |
| Consensus MFE | -26.82 |
| Energy contribution | -26.94 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.913759 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15166217 106 + 20766785 UUCAUACGACAGUCAAAUAAACGCGGACUGCAGCCACAAAGAAAUGUGUGUGUAUCCUUAUCACACAGCACAUAAAUAAUAUUUAUUGUUCUUUAUGGGGCAG------------CGA .....................((((((.((((..((((......))))..)))))))..........((.((((((.((((.....)))).))))))..)).)------------)). ( -23.60) >DroSec_CAF1 4586 118 + 1 UUCAUACGACAGUCAAAUAAACGCGGACUGCAGCCACAAAGAAAUGUGUGUGUAUCCUUAUCACACAGCACAUAAAUAAUAUUUAUUGUUCUUUAUGGGGCAGCUCAAAGGGCCGCGA .....................(((((.((((..(((.((((((((((((((((.........))))).))))).((((.....)))).)))))).))).))))((.....))))))). ( -30.50) >DroSim_CAF1 6239 118 + 1 UUCAUACGACAGUCAAAUAAACGCGGACUGCAGCCACAAAGAAAUGUGUGUGUAUCCUUAUCACACAGCACAUAAAUAAUAUUUAUUGUUCUUUAUGGGGCAGCUCAAAGGGCCGCGA .....................(((((.((((..(((.((((((((((((((((.........))))).))))).((((.....)))).)))))).))).))))((.....))))))). ( -30.50) >DroEre_CAF1 6411 118 + 1 CUCAUACGGCAGUCAAAUAAGCGCGGACUGCAGCCGCAAAGGAAUGUGUGUGUAUCCUUAUCACACAGCACAUAAAUAAUAUUUAUUGUUCUUUAUGGGGCAGCACAAAGGGCCGCGA ......((((((((...........))))((.(((.(((((((((((((((((.........))))).)))((((((...)))))).))))))).)).))).)).......))))... ( -30.70) >DroYak_CAF1 4604 118 + 1 UUCAUACGACAGUCAAAUAAACGCGGACUGCAGCCGCAAAGGAAUGUGUGUGUAUCCUUAUCACACAGCACAUAAAUAAUAUUUAUUGUUCUUUAUGGGGCAGCUCGAAGGGCCGCGA .....................(((((...((.(((.(((((((((((((((((.........))))).)))((((((...)))))).))))))).)).))).))((....))))))). ( -31.00) >consensus UUCAUACGACAGUCAAAUAAACGCGGACUGCAGCCACAAAGAAAUGUGUGUGUAUCCUUAUCACACAGCACAUAAAUAAUAUUUAUUGUUCUUUAUGGGGCAGCUCAAAGGGCCGCGA .....................(((((.((((..(((.((((((((((((((((.........))))).))))).((((.....)))).)))))).))).)))).........))))). (-26.82 = -26.94 + 0.12)

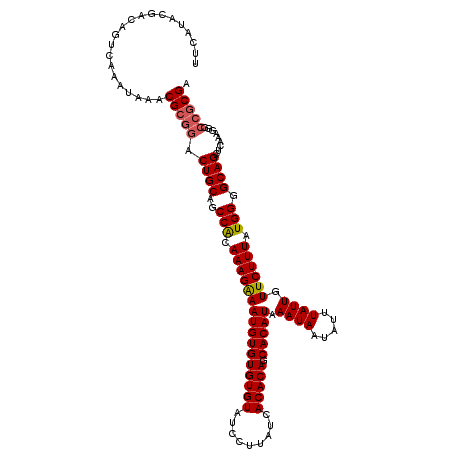
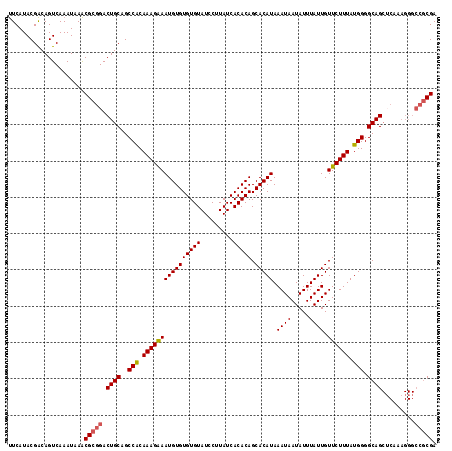
| Location | 15,166,217 – 15,166,323 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 93.39 |
| Mean single sequence MFE | -30.12 |
| Consensus MFE | -26.96 |
| Energy contribution | -28.24 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.02 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.781320 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15166217 106 - 20766785 UCG------------CUGCCCCAUAAAGAACAAUAAAUAUUAUUUAUGUGCUGUGUGAUAAGGAUACACACACAUUUCUUUGUGGCUGCAGUCCGCGUUUAUUUGACUGUCGUAUGAA ..(------------((((.((((((((((..((((((...))))))(((.((((((.......)))))))))..))))))))))..)))))....(((.....)))........... ( -26.10) >DroSec_CAF1 4586 118 - 1 UCGCGGCCCUUUGAGCUGCCCCAUAAAGAACAAUAAAUAUUAUUUAUGUGCUGUGUGAUAAGGAUACACACACAUUUCUUUGUGGCUGCAGUCCGCGUUUAUUUGACUGUCGUAUGAA .(((((........(((((.((((((((((..((((((...))))))(((.((((((.......)))))))))..))))))))))..))))))))))(((((.((.....)).))))) ( -33.20) >DroSim_CAF1 6239 118 - 1 UCGCGGCCCUUUGAGCUGCCCCAUAAAGAACAAUAAAUAUUAUUUAUGUGCUGUGUGAUAAGGAUACACACACAUUUCUUUGUGGCUGCAGUCCGCGUUUAUUUGACUGUCGUAUGAA .(((((........(((((.((((((((((..((((((...))))))(((.((((((.......)))))))))..))))))))))..))))))))))(((((.((.....)).))))) ( -33.20) >DroEre_CAF1 6411 118 - 1 UCGCGGCCCUUUGUGCUGCCCCAUAAAGAACAAUAAAUAUUAUUUAUGUGCUGUGUGAUAAGGAUACACACACAUUCCUUUGCGGCUGCAGUCCGCGCUUAUUUGACUGCCGUAUGAG ..(((((.(((((((......))))))).....((((((......(((((.((((((.......)))))))))))......(((((....).))))...))))))...)))))..... ( -30.60) >DroYak_CAF1 4604 118 - 1 UCGCGGCCCUUCGAGCUGCCCCAUAAAGAACAAUAAAUAUUAUUUAUGUGCUGUGUGAUAAGGAUACACACACAUUCCUUUGCGGCUGCAGUCCGCGUUUAUUUGACUGUCGUAUGAA ..(((((....((..((((.((.(((((....((((((...))))))(((.((((((.......)))))))))....))))).))..))))..)).(((.....))).)))))..... ( -27.50) >consensus UCGCGGCCCUUUGAGCUGCCCCAUAAAGAACAAUAAAUAUUAUUUAUGUGCUGUGUGAUAAGGAUACACACACAUUUCUUUGUGGCUGCAGUCCGCGUUUAUUUGACUGUCGUAUGAA .(((((........(((((.((((((((((..((((((...))))))(((.((((((.......)))))))))..))))))))))..))))))))))(((((.((.....)).))))) (-26.96 = -28.24 + 1.28)

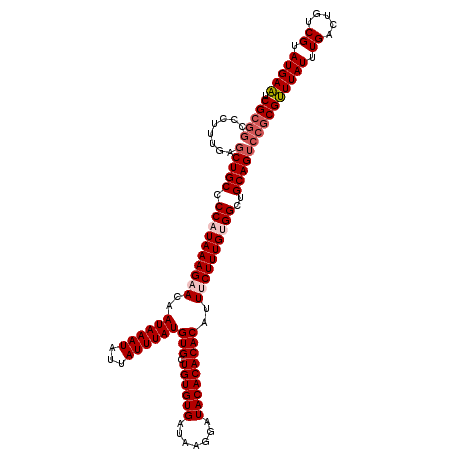
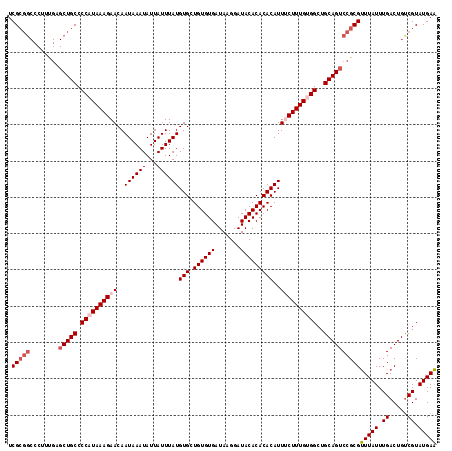
| Location | 15,166,323 – 15,166,418 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.14 |
| Mean single sequence MFE | -19.87 |
| Consensus MFE | -14.50 |
| Energy contribution | -14.67 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.877515 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15166323 95 + 20766785 AGAUGCCAACCCCACGGCCUAGUGUCAGUUGGGAAACAUUCAAGUGUUUCAAAUAUUUCUUAACAGCCCUCCCC--------------UGACACUU------GUUUACACACACA----- ....(((........)))..((((((((..(((((((((....)))))))...((.....)).....))....)--------------))))))).------.............----- ( -20.40) >DroSec_CAF1 4704 84 + 1 AGAUGCCAACCCCACGGCCUAGUGUCAGUUGGGAAACAUUCAAGUGUUUCAAAUAUUUUUUAACAGCUUUCCCC--------------CGACACUU------GU---------------- ....(((........)))..(((((((((((.(((((((....)))))))(((.....)))..)))))......--------------.)))))).------..---------------- ( -18.01) >DroSim_CAF1 6357 84 + 1 AGAUGCCAACCCCACGGCCUAGUGUCAGUUGGGAAACAUUCAAGUGUUUCAAAUAUUUUUUAACAGCUUUCCCC--------------CGACACUU------GU---------------- ....(((........)))..(((((((((((.(((((((....)))))))(((.....)))..)))))......--------------.)))))).------..---------------- ( -18.01) >DroEre_CAF1 6529 75 + 1 AGAUGCCAACCCCACGGCCUAGUGUCAGUUGGGAAACAUUCAAGUGUUUCAAAUAUUUUUUAACAGCUUUCCCCC--------------------------------------------- .....(((((..(((......)))...)))))(((((((....))))))).........................--------------------------------------------- ( -15.40) >DroYak_CAF1 4722 102 + 1 AGAUGCCAACCCCACGGCCUAGUGUCAGUUGGGAAACAUUCAAGUGUUUCAAAUAUUUUUUAACAGCUUUCCCCC-------------CGACACUUAUACGUGUAUACACACCCA----- ............((((....(((((((((((.(((((((....)))))))(((.....)))..))))).......-------------.))))))....))))............----- ( -21.10) >DroAna_CAF1 4309 110 + 1 AGAUGCCAACCCCAAGGCCUAGUGUCAGUUGGGAAACAUUCAAGUGUUUCGAAUAUUUUUUAACAGCUUCCCCCCACGGUGGAAAGCCUGCCACUC------C----CACUCCCACUCCU ....(((........)))..((((..(((.(((((((((....))))))).............((((((((((....)).)).))).))).....)------)----.)))..))))... ( -26.30) >consensus AGAUGCCAACCCCACGGCCUAGUGUCAGUUGGGAAACAUUCAAGUGUUUCAAAUAUUUUUUAACAGCUUUCCCC______________CGACACUU______GU________________ .....(((((..(((......)))...)))))(((((((....)))))))...................................................................... (-14.50 = -14.67 + 0.17)

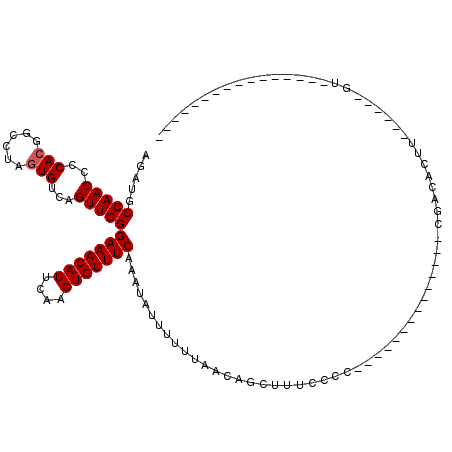
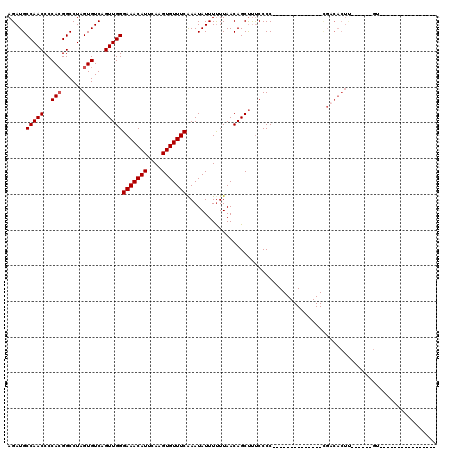
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:25:26 2006