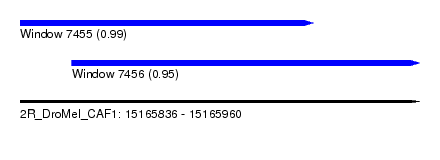
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 15,165,836 – 15,165,960 |
| Length | 124 |
| Max. P | 0.993969 |

| Location | 15,165,836 – 15,165,927 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 80.97 |
| Mean single sequence MFE | -32.27 |
| Consensus MFE | -18.98 |
| Energy contribution | -19.73 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.36 |
| Structure conservation index | 0.59 |
| SVM decision value | 2.44 |
| SVM RNA-class probability | 0.993969 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15165836 91 + 20766785 AUA---CAUACAUAUCAAGUGACACAGGUGCUUGAAAUGGUGGGCUUAAAGGCUCAUUUAAGCCCUCUUUGUGUGUG-----UCAUUGGGCCAACUCAC ..(---((((((((((((((.((....))))))))...((.(((((((((......))))))))).)).))))))))-----)...((((....)))). ( -30.10) >DroSec_CAF1 4262 94 + 1 GCA---CAUACAUAUCAAGUGACACAGGUGCUUGAAAUGGUGGGCUUAAAGGCUCAUUUAAGCCCUCUCCGUGUGUGUGCGCUCUUUGGGCCAACUA-- (((---(((((((.((((((.((....))))))))...((.(((((((((......)))))))))...))))))))))))(((.....)))......-- ( -39.50) >DroEre_CAF1 6053 88 + 1 AUACUAUGUACAUAUCAUGUGACACAGGUGCUGGAAAUGGUGGGCUUAAAGGCUCAUUUAAGCCCUCAGUGUAUGUGUGCG---------CCAACUA-- ..................(((.((((.((((......(((.(((((((((......))))))))))))..)))).))))))---------)......-- ( -26.50) >DroYak_CAF1 4232 90 + 1 AU-------ACAUAUCAUGUGACACAGGUGCUUGAAAUGGAGGGCUUAAAGGCUCAUUUAAGCCCUCUUUGCAUGUGUGCGCUCAUUGGGCCAACUA-- ..-------.....(((.(((.((((.((((.......((((((((((((......))))))))))))..)))).)))))))....)))........-- ( -33.00) >consensus AUA___CAUACAUAUCAAGUGACACAGGUGCUUGAAAUGGUGGGCUUAAAGGCUCAUUUAAGCCCUCUUUGUAUGUGUGCG_UCAUUGGGCCAACUA__ ..........(((.((((((.((....)))))))).)))(.(((((((((......))))))))).)................................ (-18.98 = -19.73 + 0.75)

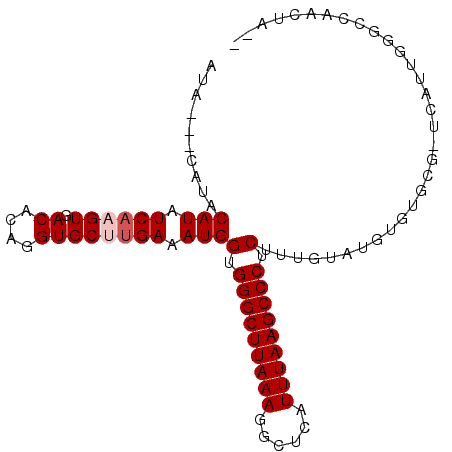
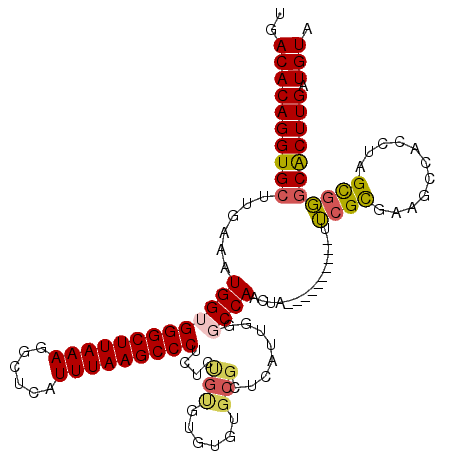
| Location | 15,165,852 – 15,165,960 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.70 |
| Mean single sequence MFE | -40.32 |
| Consensus MFE | -27.80 |
| Energy contribution | -28.04 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.949175 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15165852 108 + 20766785 UGACACAGGUGCUUGAAAUGGUGGGCUUAAAGGCUCAUUUAAGCCCUCUUUGUGUGUG-----UCAUUGGGCCAACUCACUAGGUGGCUGCGCGAA------UAGU-UGCACUUGAUGUA ..((((((((((.......((.(((((((((......))))))))).))(((((((.(-----((((..((........))..)))))))))))).------....-.))))))).))). ( -38.40) >DroSec_CAF1 4278 110 + 1 UGACACAGGUGCUUGAAAUGGUGGGCUUAAAGGCUCAUUUAAGCCCUCUCCGUGUGUGUGCGCUCUUUGGGCCAACUA----------UUCGCGAAGCCACCUAGCGGGCACUUGAUGUA ..(((((((((((((...(((((((((((((......)))))))))....((((....((.(((.....)))))....----------..))))..)))).....)))))))))).))). ( -42.20) >DroSim_CAF1 5888 110 + 1 UGACACAGGUGCUUGAAAUGGUGGGCUUAAAGGCUCAUUUAAGCCCUCUCCGUGUGUGUGCGCUCAUUGGGCCAACUA----------UUCGCGAAGCCACCUAGCGGGCACUUGAUGUA ..(((((((((((((...(((((((((((((......)))))))))....((((....((.(((.....)))))....----------..))))..)))).....)))))))))).))). ( -42.20) >DroEre_CAF1 6072 101 + 1 UGACACAGGUGCUGGAAAUGGUGGGCUUAAAGGCUCAUUUAAGCCCUCAGUGUAUGUGUGCG---------CCAACUA----------UUCGUGCAGCCACCUAGCGGGCGCUUGAUGUA ..(((((((((((.(...(((((((((((((......)))))))))....((((((.((...---------...))..----------..)))))))))).....).)))))))).))). ( -37.60) >DroYak_CAF1 4244 110 + 1 UGACACAGGUGCUUGAAAUGGAGGGCUUAAAGGCUCAUUUAAGCCCUCUUUGCAUGUGUGCGCUCAUUGGGCCAACUA----------UUCGUGCAGCCACCUAGCGGCCACUUGAUGUA .(.((((.((((.......((((((((((((......))))))))))))..)))).)))))(((((...((((..(((----------...(((....))).))).))))...))).)). ( -41.20) >consensus UGACACAGGUGCUUGAAAUGGUGGGCUUAAAGGCUCAUUUAAGCCCUCUCUGUGUGUGUGCGCUCAUUGGGCCAACUA__________UUCGCGAAGCCACCUAGCGGGCACUUGAUGUA ..((((((((((......(((((((((((((......)))))))))....(((......)))........))))...............((((...........))))))))))).))). (-27.80 = -28.04 + 0.24)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:25:24 2006