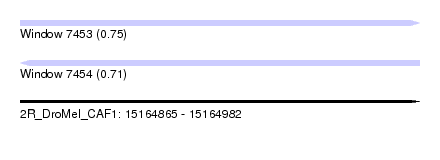
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 15,164,865 – 15,164,982 |
| Length | 117 |
| Max. P | 0.753095 |

| Location | 15,164,865 – 15,164,982 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 91.37 |
| Mean single sequence MFE | -27.62 |
| Consensus MFE | -20.57 |
| Energy contribution | -21.37 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.753095 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
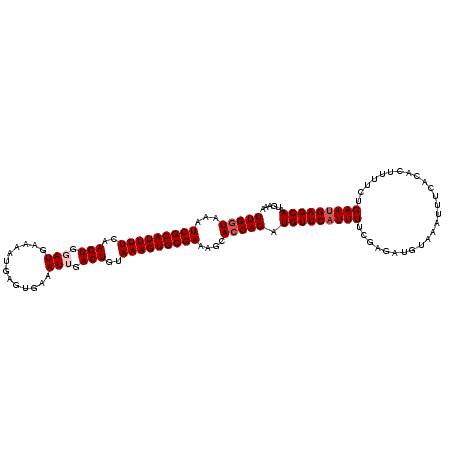
>2R_DroMel_CAF1 15164865 117 + 20766785 UUGGUAAAUCGCAUUUUCAAGUGGAUGAAAAUGAGUGAAAUUGACUGUAAAGUGCGAAAGCACCAGAUGUUCAUUUGCGAGAUGUAAAUUUCACGCUUUUCUGAAUGAGCAAUGAAA (((((...(((((((((..(((.(((.............))).)))..)))))))))....))))).((((((((((((.((........)).)))......)))))))))...... ( -29.82) >DroSec_CAF1 3273 105 + 1 UUGGUAAAUCGCAUUUUCAAGUGGAUGAAAAUGAGUGAAAUUGACUGUAAAGUGCGAAAGAACCAGAUGUUCAUUUU------------UUCACACUUUUCUGAAUGAGCAAUGAAA (((((...(((((((((..(((.(((.............))).)))..)))))))))....))))).(((((((((.------------.............)))))))))...... ( -25.36) >DroSim_CAF1 3272 117 + 1 UUGGUAAAUCGCAUUUUCAAGUGGAUGAAAAUGAGUGAAAUUGACUGUAAAGUGCGAAAGCACCAGAUGUUCAUUUUCGAGAUGUAAAUUUCACACUUUUCUGAAUGAGCAAUGAAA (((((...(((((((((..(((.(((.............))).)))..)))))))))....))))).(((((((((..(((((((.......))).))))..)))))))))...... ( -28.72) >DroEre_CAF1 3693 117 + 1 UUGGUAAAUCGCAUUUUCAAGUGGAUGAAAAUGAGUGAAAUGGACUGUAAAGUGCGAAAGCACCAGAUGUUCAUUUUCCAGAUGUAAAUUUUACACUCUUCUGAACGAGCAAGGAAA ((((((((...((((((((......)))))))).(..((((((((......((((....)))).....))))))))..)..........))))).(((........))))))..... ( -28.40) >DroYak_CAF1 3217 117 + 1 UUGGUAAAUCGCAUUUUCAAGUGGAUAAAAAUGAGUGAAAUGGACUGUAAAGUGCGAAAGCAGCAGAUGUUCAUUUGCGAGAUGUAAAUUUCACACUAUUCUGAAUGAGCAACGAAA ..........((...(((((((((((....))..(((((((..((.......(((....)))((((((....)))))).....))..))))))).))))).))))...))....... ( -25.80) >consensus UUGGUAAAUCGCAUUUUCAAGUGGAUGAAAAUGAGUGAAAUUGACUGUAAAGUGCGAAAGCACCAGAUGUUCAUUUUCGAGAUGUAAAUUUCACACUUUUCUGAAUGAGCAAUGAAA (((((...(((((((((..(((.(((.............))).)))..)))))))))....))))).(((((((((..........................)))))))))...... (-20.57 = -21.37 + 0.80)


| Location | 15,164,865 – 15,164,982 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 91.37 |
| Mean single sequence MFE | -22.30 |
| Consensus MFE | -15.94 |
| Energy contribution | -17.02 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.713462 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
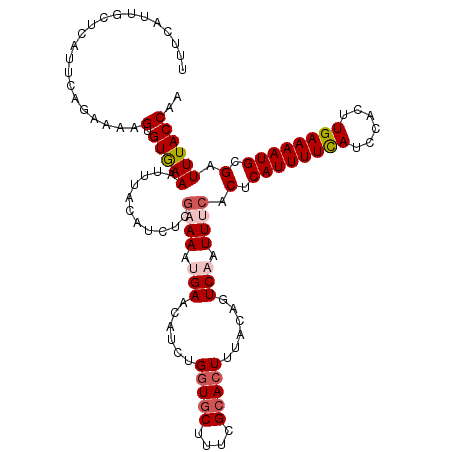
>2R_DroMel_CAF1 15164865 117 - 20766785 UUUCAUUGCUCAUUCAGAAAAGCGUGAAAUUUACAUCUCGCAAAUGAACAUCUGGUGCUUUCGCACUUUACAGUCAAUUUCACUCAUUUUCAUCCACUUGAAAAUGCGAUUUACCAA ....(((((.......((((.(((.((........)).)))...(((......(((((....)))))......))).))))....(((((((......))))))))))))....... ( -23.80) >DroSec_CAF1 3273 105 - 1 UUUCAUUGCUCAUUCAGAAAAGUGUGAA------------AAAAUGAACAUCUGGUUCUUUCGCACUUUACAGUCAAUUUCACUCAUUUUCAUCCACUUGAAAAUGCGAUUUACCAA ....(((((.......(.((((((((((------------(....((((.....))))))))))))))).)..............(((((((......))))))))))))....... ( -22.50) >DroSim_CAF1 3272 117 - 1 UUUCAUUGCUCAUUCAGAAAAGUGUGAAAUUUACAUCUCGAAAAUGAACAUCUGGUGCUUUCGCACUUUACAGUCAAUUUCACUCAUUUUCAUCCACUUGAAAAUGCGAUUUACCAA ....(((((............(.((((((((.((.((........))......(((((....))))).....)).)))))))).)(((((((......))))))))))))....... ( -23.50) >DroEre_CAF1 3693 117 - 1 UUUCCUUGCUCGUUCAGAAGAGUGUAAAAUUUACAUCUGGAAAAUGAACAUCUGGUGCUUUCGCACUUUACAGUCCAUUUCACUCAUUUUCAUCCACUUGAAAAUGCGAUUUACCAA .......((((........))))..............(((.(((((.((....(((((....))))).....)).)))))..(.((((((((......)))))))).).....))). ( -23.20) >DroYak_CAF1 3217 117 - 1 UUUCGUUGCUCAUUCAGAAUAGUGUGAAAUUUACAUCUCGCAAAUGAACAUCUGCUGCUUUCGCACUUUACAGUCCAUUUCACUCAUUUUUAUCCACUUGAAAAUGCGAUUUACCAA ....(((((............(.(((((((..((.....(((.((....)).)))(((....))).......))..))))))).)(((((((......))))))))))))....... ( -18.50) >consensus UUUCAUUGCUCAUUCAGAAAAGUGUGAAAUUUACAUCUCGAAAAUGAACAUCUGGUGCUUUCGCACUUUACAGUCAAUUUCACUCAUUUUCAUCCACUUGAAAAUGCGAUUUACCAA .....................(.(((((...........((((.(((......(((((....)))))......))).)))).(.((((((((......)))))))).).)))))).. (-15.94 = -17.02 + 1.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:25:22 2006