
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 15,163,882 – 15,164,049 |
| Length | 167 |
| Max. P | 0.999432 |

| Location | 15,163,882 – 15,163,988 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 88.29 |
| Mean single sequence MFE | -26.72 |
| Consensus MFE | -22.06 |
| Energy contribution | -22.45 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.868082 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
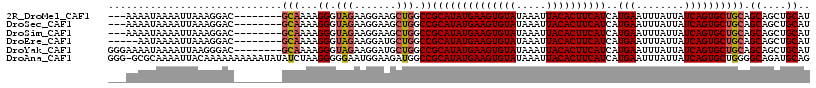
>2R_DroMel_CAF1 15163882 106 + 20766785 ---AAAAUAAAAUUAAAGGAC--------GCAAAAGGGUAGAAGGAAGCUGGCCGCAUAUGAAGUGUAUAAAUUACACUUCAUCAUGAAUUUAUUAUCAGUGCUGCAGCAGCUGCAU ---..((((((..........--------.......(((...((....)).))).(((((((((((((.....)))))))))).)))..))))))....((((.((....)).)))) ( -26.10) >DroSec_CAF1 2283 106 + 1 ---AAAAUAAAAUUAAAGGAC--------GCAAAAGGGUAGAAGGAAGCUGGCCGCAUAUGAAGUGUAUAAAUUACACUUCAUCAUGAAUUUAUUAUCAGUGCUGCAGCAGCUGCAU ---..((((((..........--------.......(((...((....)).))).(((((((((((((.....)))))))))).)))..))))))....((((.((....)).)))) ( -26.10) >DroSim_CAF1 2290 106 + 1 ---AAAAUAAAAUUAAAGGAC--------GCAAAAGGGUAGAAGGAAGCUGGCCGCAUAUGAAGUGUAUAAAUUACACUUCAUCAUGAAUUUAUUAUCAGUGCUGCAGCAGCUGCAU ---..((((((..........--------.......(((...((....)).))).(((((((((((((.....)))))))))).)))..))))))....((((.((....)).)))) ( -26.10) >DroEre_CAF1 2626 104 + 1 -----AAUAAAAUUAAAGGAC--------GCAAAAGGGUAGAAGGAUGCUGGCCGCAUAUGAAGUGUAUAAAUUACACUUCAUCAUGAAUUUAUUAUCAGUGCUGCAGCAGCUGCAU -----................--------........((((.....(((((.(.((((((((((((((.....))))))))))..(((........))))))).)))))).)))).. ( -28.00) >DroYak_CAF1 2264 109 + 1 GGGAAAAUAAAAUUAAGGGAC--------GCAAAAGGGUAGAAGGAUGCUGGCCGCAUAUGAAGUGUAUAAAUUACACUUCAUCAUGAAUUUAUUAUCAGUGCUGCAGCAGCUGCAU .....................--------........((((.....(((((.(.((((((((((((((.....))))))))))..(((........))))))).)))))).)))).. ( -28.00) >DroAna_CAF1 2214 116 + 1 GGG-GCGCAAAAUUACAAAAAAAAAAUAUAUCUAAGGGGGAAUGGAAGAUGGCCGCAUAUGAAGUGUAUAAAUUACACUUCAUCAUGAAUUUAUUAUCAGUGCUGGGGCAGAUGCAG ..(-((((......................(((.....)))...((.(((((...(((((((((((((.....)))))))))).)))...))))).)).)))))...((....)).. ( -26.00) >consensus ___AAAAUAAAAUUAAAGGAC________GCAAAAGGGUAGAAGGAAGCUGGCCGCAUAUGAAGUGUAUAAAUUACACUUCAUCAUGAAUUUAUUAUCAGUGCUGCAGCAGCUGCAU .............................(((...((.(((.......))).))((((((((((((((.....))))))))))..(((........)))))))))).((....)).. (-22.06 = -22.45 + 0.39)
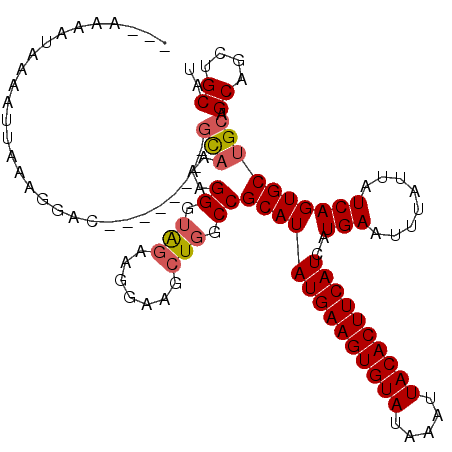
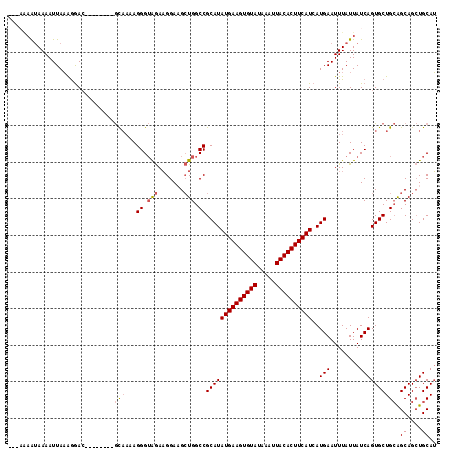
| Location | 15,163,882 – 15,163,988 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 88.29 |
| Mean single sequence MFE | -25.64 |
| Consensus MFE | -23.97 |
| Energy contribution | -24.33 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.33 |
| Structure conservation index | 0.94 |
| SVM decision value | 3.60 |
| SVM RNA-class probability | 0.999432 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15163882 106 - 20766785 AUGCAGCUGCUGCAGCACUGAUAAUAAAUUCAUGAUGAAGUGUAAUUUAUACACUUCAUAUGCGGCCAGCUUCCUUCUACCCUUUUGC--------GUCCUUUAAUUUUAUUUU--- ((((((((((((((.((.(((........)))))((((((((((.....)))))))))).))))).))))...............)))--------))................--- ( -26.66) >DroSec_CAF1 2283 106 - 1 AUGCAGCUGCUGCAGCACUGAUAAUAAAUUCAUGAUGAAGUGUAAUUUAUACACUUCAUAUGCGGCCAGCUUCCUUCUACCCUUUUGC--------GUCCUUUAAUUUUAUUUU--- ((((((((((((((.((.(((........)))))((((((((((.....)))))))))).))))).))))...............)))--------))................--- ( -26.66) >DroSim_CAF1 2290 106 - 1 AUGCAGCUGCUGCAGCACUGAUAAUAAAUUCAUGAUGAAGUGUAAUUUAUACACUUCAUAUGCGGCCAGCUUCCUUCUACCCUUUUGC--------GUCCUUUAAUUUUAUUUU--- ((((((((((((((.((.(((........)))))((((((((((.....)))))))))).))))).))))...............)))--------))................--- ( -26.66) >DroEre_CAF1 2626 104 - 1 AUGCAGCUGCUGCAGCACUGAUAAUAAAUUCAUGAUGAAGUGUAAUUUAUACACUUCAUAUGCGGCCAGCAUCCUUCUACCCUUUUGC--------GUCCUUUAAUUUUAUU----- ((((((((((((((.((.(((........)))))((((((((((.....)))))))))).))))).))))...............)))--------))..............----- ( -26.66) >DroYak_CAF1 2264 109 - 1 AUGCAGCUGCUGCAGCACUGAUAAUAAAUUCAUGAUGAAGUGUAAUUUAUACACUUCAUAUGCGGCCAGCAUCCUUCUACCCUUUUGC--------GUCCCUUAAUUUUAUUUUCCC ((((((((((((((.((.(((........)))))((((((((((.....)))))))))).))))).))))...............)))--------))................... ( -26.66) >DroAna_CAF1 2214 116 - 1 CUGCAUCUGCCCCAGCACUGAUAAUAAAUUCAUGAUGAAGUGUAAUUUAUACACUUCAUAUGCGGCCAUCUUCCAUUCCCCCUUAGAUAUAUUUUUUUUUUGUAAUUUUGCGC-CCC ..(((...(((.((.((.(((........)))))((((((((((.....)))))))))).)).))).((((.............))))....................)))..-... ( -20.52) >consensus AUGCAGCUGCUGCAGCACUGAUAAUAAAUUCAUGAUGAAGUGUAAUUUAUACACUUCAUAUGCGGCCAGCUUCCUUCUACCCUUUUGC________GUCCUUUAAUUUUAUUUU___ .....(((((((((.((.(((........)))))((((((((((.....)))))))))).))))).))))............................................... (-23.97 = -24.33 + 0.36)

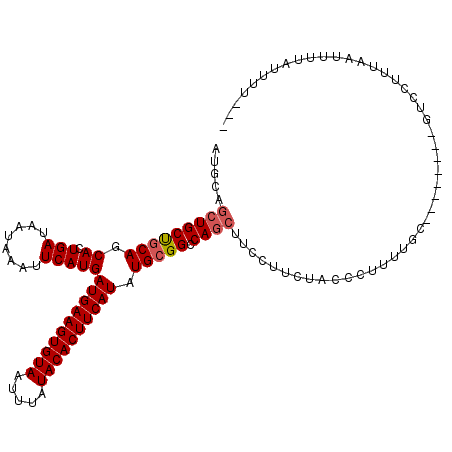
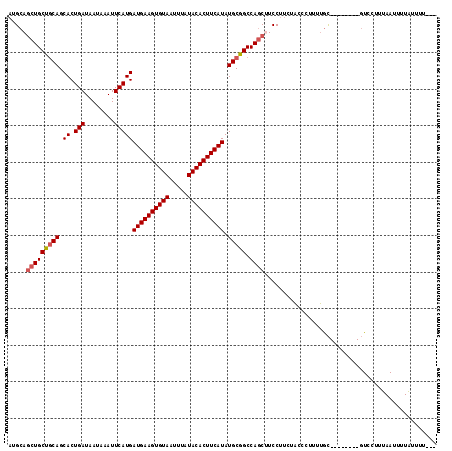
| Location | 15,163,911 – 15,164,009 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 90.86 |
| Mean single sequence MFE | -33.18 |
| Consensus MFE | -26.97 |
| Energy contribution | -27.30 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.95 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.925914 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15163911 98 + 20766785 GAAGGAAGCUGGCCGCAUAUGAAGUGUAUAAAUUACACUUCAUCAUGAAUUUAUUAUCAGUGCUGCAGCAGCUGCAUCUGGAG------------------CUGCCACGA-AUGCAC .......((..((.((((((((((((((.....))))))))))..(((........))))))).)).((((((.(....).))------------------)))).....-..)).. ( -32.20) >DroSec_CAF1 2312 98 + 1 GAAGGAAGCUGGCCGCAUAUGAAGUGUAUAAAUUACACUUCAUCAUGAAUUUAUUAUCAGUGCUGCAGCAGCUGCAUCUGGAG------------------CUGCCACGA-AUGCAC .......((..((.((((((((((((((.....))))))))))..(((........))))))).)).((((((.(....).))------------------)))).....-..)).. ( -32.20) >DroSim_CAF1 2319 98 + 1 GAAGGAAGCUGGCCGCAUAUGAAGUGUAUAAAUUACACUUCAUCAUGAAUUUAUUAUCAGUGCUGCAGCAGCUGCAUCUGGAG------------------CUGCCACGA-AUGCAC .......((..((.((((((((((((((.....))))))))))..(((........))))))).)).((((((.(....).))------------------)))).....-..)).. ( -32.20) >DroEre_CAF1 2653 98 + 1 GAAGGAUGCUGGCCGCAUAUGAAGUGUAUAAAUUACACUUCAUCAUGAAUUUAUUAUCAGUGCUGCAGCAGCUGCAUCUGGAG------------------CUGCCACGA-AUGCUC .......((..((.((((((((((((((.....))))))))))..(((........))))))).)).((((((.(....).))------------------)))).....-..)).. ( -31.50) >DroYak_CAF1 2296 98 + 1 GAAGGAUGCUGGCCGCAUAUGAAGUGUAUAAAUUACACUUCAUCAUGAAUUUAUUAUCAGUGCUGCAGCAGCUGCAUCUGGAG------------------CUGCCACGA-AUGCAC ......(((..((.((((((((((((((.....))))))))))..(((........))))))).)).((((((.(....).))------------------)))).....-..))). ( -32.70) >DroAna_CAF1 2253 117 + 1 AAUGGAAGAUGGCCGCAUAUGAAGUGUAUAAAUUACACUUCAUCAUGAAUUUAUUAUCAGUGCUGGGGCAGAUGCAGCUGGAGGUGAAGCUGAUGCUGAUGCUGCCACGAAAUGCAC ....((.(((((...(((((((((((((.....)))))))))).)))...))))).)).((((...(((((...((((....((.....))...))))...))))).......)))) ( -38.30) >consensus GAAGGAAGCUGGCCGCAUAUGAAGUGUAUAAAUUACACUUCAUCAUGAAUUUAUUAUCAGUGCUGCAGCAGCUGCAUCUGGAG__________________CUGCCACGA_AUGCAC .......(.((((..(((((((((((((.....)))))))))).))).........(((((((.((....)).))).))))......................)))))......... (-26.97 = -27.30 + 0.33)

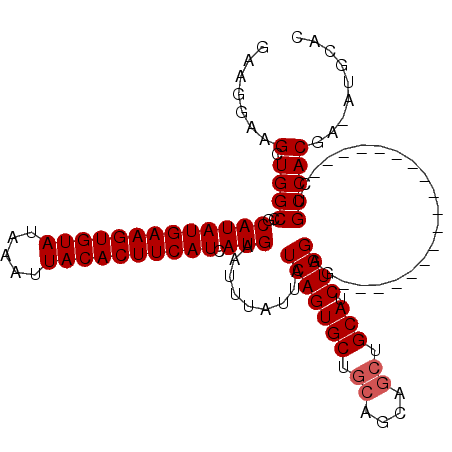
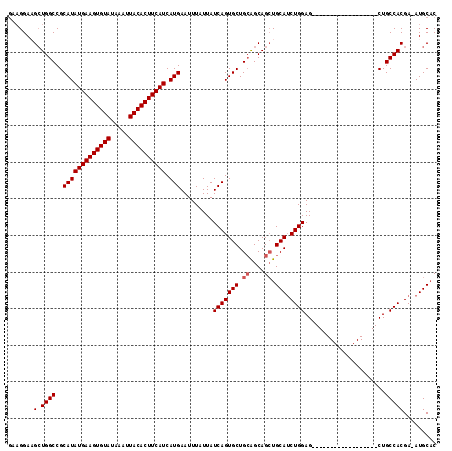
| Location | 15,163,911 – 15,164,009 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 90.86 |
| Mean single sequence MFE | -32.90 |
| Consensus MFE | -27.04 |
| Energy contribution | -27.74 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.53 |
| Structure conservation index | 0.82 |
| SVM decision value | 2.00 |
| SVM RNA-class probability | 0.985330 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15163911 98 - 20766785 GUGCAU-UCGUGGCAG------------------CUCCAGAUGCAGCUGCUGCAGCACUGAUAAUAAAUUCAUGAUGAAGUGUAAUUUAUACACUUCAUAUGCGGCCAGCUUCCUUC .(((((-(..(((...------------------..)))))))))(((((((((.((.(((........)))))((((((((((.....)))))))))).))))).))))....... ( -33.00) >DroSec_CAF1 2312 98 - 1 GUGCAU-UCGUGGCAG------------------CUCCAGAUGCAGCUGCUGCAGCACUGAUAAUAAAUUCAUGAUGAAGUGUAAUUUAUACACUUCAUAUGCGGCCAGCUUCCUUC .(((((-(..(((...------------------..)))))))))(((((((((.((.(((........)))))((((((((((.....)))))))))).))))).))))....... ( -33.00) >DroSim_CAF1 2319 98 - 1 GUGCAU-UCGUGGCAG------------------CUCCAGAUGCAGCUGCUGCAGCACUGAUAAUAAAUUCAUGAUGAAGUGUAAUUUAUACACUUCAUAUGCGGCCAGCUUCCUUC .(((((-(..(((...------------------..)))))))))(((((((((.((.(((........)))))((((((((((.....)))))))))).))))).))))....... ( -33.00) >DroEre_CAF1 2653 98 - 1 GAGCAU-UCGUGGCAG------------------CUCCAGAUGCAGCUGCUGCAGCACUGAUAAUAAAUUCAUGAUGAAGUGUAAUUUAUACACUUCAUAUGCGGCCAGCAUCCUUC ..((((-(..(((...------------------..)))))))).(((((((((.((.(((........)))))((((((((((.....)))))))))).))))).))))....... ( -32.80) >DroYak_CAF1 2296 98 - 1 GUGCAU-UCGUGGCAG------------------CUCCAGAUGCAGCUGCUGCAGCACUGAUAAUAAAUUCAUGAUGAAGUGUAAUUUAUACACUUCAUAUGCGGCCAGCAUCCUUC .(((((-(..(((...------------------..)))))))))(((((((((.((.(((........)))))((((((((((.....)))))))))).))))).))))....... ( -33.50) >DroAna_CAF1 2253 117 - 1 GUGCAUUUCGUGGCAGCAUCAGCAUCAGCUUCACCUCCAGCUGCAUCUGCCCCAGCACUGAUAAUAAAUUCAUGAUGAAGUGUAAUUUAUACACUUCAUAUGCGGCCAUCUUCCAUU .........(((((.(((((((...(((((........)))))....(((....))))))))............((((((((((.....))))))))))..)).)))))........ ( -32.10) >consensus GUGCAU_UCGUGGCAG__________________CUCCAGAUGCAGCUGCUGCAGCACUGAUAAUAAAUUCAUGAUGAAGUGUAAUUUAUACACUUCAUAUGCGGCCAGCUUCCUUC .(((((....(((.((..................))))).)))))(((((((((.((.(((........)))))((((((((((.....)))))))))).))))).))))....... (-27.04 = -27.74 + 0.70)

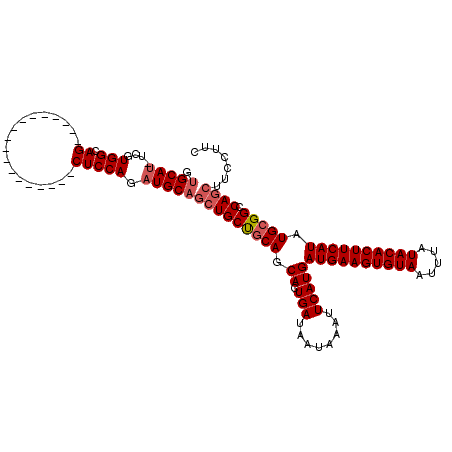
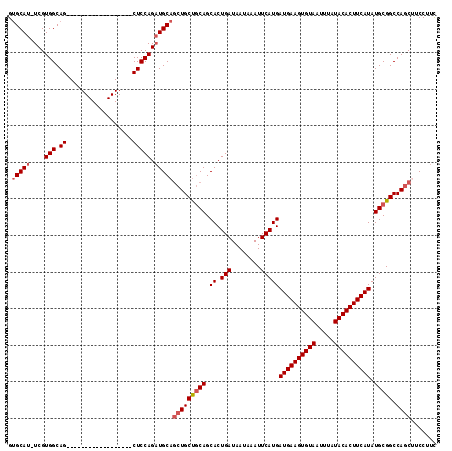
| Location | 15,163,948 – 15,164,049 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.75 |
| Mean single sequence MFE | -36.52 |
| Consensus MFE | -27.28 |
| Energy contribution | -28.00 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.07 |
| Structure conservation index | 0.75 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.529673 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15163948 101 + 20766785 CUUCAUCAUGAAUUUAUUAUCAGUGCUGCAGCAGCUGCAUCUGGAG------------------CUGCCACGA-AUGCACCAUUGCAGCACGCUGGCUCAGCACAAGAUGUGCUCCUUCC ...((((.(((........)))((((((((((.((((((..(((.(------------------(........-..)).))).))))))..))))...))))))..)))).......... ( -33.20) >DroSec_CAF1 2349 101 + 1 CUUCAUCAUGAAUUUAUUAUCAGUGCUGCAGCAGCUGCAUCUGGAG------------------CUGCCACGA-AUGCACCACUGCAGCACGCUGGCUCAGCACAAGAUGUGCUCCUUCC ...((((.((............(((((((((..(.(((((((((..------------------...))).).-))))))..)))))))))((((...)))).)).)))).......... ( -33.70) >DroSim_CAF1 2356 101 + 1 CUUCAUCAUGAAUUUAUUAUCAGUGCUGCAGCAGCUGCAUCUGGAG------------------CUGCCACGA-AUGCACCACUGCAGCACGCUGGCUCAGCACAAGAUGUGCUCCUUCC ...((((.((............(((((((((..(.(((((((((..------------------...))).).-))))))..)))))))))((((...)))).)).)))).......... ( -33.70) >DroEre_CAF1 2690 101 + 1 CUUCAUCAUGAAUUUAUUAUCAGUGCUGCAGCAGCUGCAUCUGGAG------------------CUGCCACGA-AUGCUCCACUGCAGCACGCUGGCUCAGCACAAGAUGUGCUCCUUCC ...((((.(((........)))((((((((((.((((((..(((((------------------(........-..)))))).))))))..))))...))))))..)))).......... ( -37.90) >DroYak_CAF1 2333 101 + 1 CUUCAUCAUGAAUUUAUUAUCAGUGCUGCAGCAGCUGCAUCUGGAG------------------CUGCCACGA-AUGCACCACUGCAGCACGCUGGCUCAGCACAAGAUGUGCUCCUUCC ...((((.((............(((((((((..(.(((((((((..------------------...))).).-))))))..)))))))))((((...)))).)).)))).......... ( -33.70) >DroAna_CAF1 2290 120 + 1 CUUCAUCAUGAAUUUAUUAUCAGUGCUGGGGCAGAUGCAGCUGGAGGUGAAGCUGAUGCUGAUGCUGCCACGAAAUGCACCAAGGCAGCACGCUGGCUCAGCACAAGAUGAGGUCCUUGC (((((((.(((........)))((((((((.(((...(((((........))))).......(((((((..............)))))))..))).))))))))..)))))))....... ( -46.94) >consensus CUUCAUCAUGAAUUUAUUAUCAGUGCUGCAGCAGCUGCAUCUGGAG__________________CUGCCACGA_AUGCACCACUGCAGCACGCUGGCUCAGCACAAGAUGUGCUCCUUCC ...((((.((............(((((((((..(.(((((.(((.......................)))....))))))..)))))))))((((...)))).)).)))).......... (-27.28 = -28.00 + 0.72)

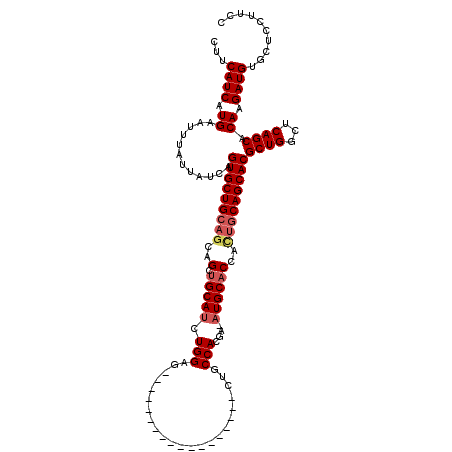
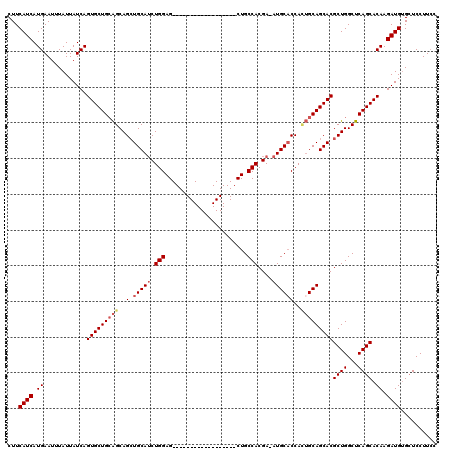
| Location | 15,163,948 – 15,164,049 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.75 |
| Mean single sequence MFE | -38.67 |
| Consensus MFE | -30.82 |
| Energy contribution | -31.82 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.744290 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15163948 101 - 20766785 GGAAGGAGCACAUCUUGUGCUGAGCCAGCGUGCUGCAAUGGUGCAU-UCGUGGCAG------------------CUCCAGAUGCAGCUGCUGCAGCACUGAUAAUAAAUUCAUGAUGAAG ..........((((..((((((...(((((.((((((.(((.((..-........)------------------).)))..)))))))))))))))))(((........))).))))... ( -35.90) >DroSec_CAF1 2349 101 - 1 GGAAGGAGCACAUCUUGUGCUGAGCCAGCGUGCUGCAGUGGUGCAU-UCGUGGCAG------------------CUCCAGAUGCAGCUGCUGCAGCACUGAUAAUAAAUUCAUGAUGAAG ((....(((((.....)))))...))...(((((((((..((((((-(..(((...------------------..))))))))).)..)))))))))(((........)))........ ( -39.20) >DroSim_CAF1 2356 101 - 1 GGAAGGAGCACAUCUUGUGCUGAGCCAGCGUGCUGCAGUGGUGCAU-UCGUGGCAG------------------CUCCAGAUGCAGCUGCUGCAGCACUGAUAAUAAAUUCAUGAUGAAG ((....(((((.....)))))...))...(((((((((..((((((-(..(((...------------------..))))))))).)..)))))))))(((........)))........ ( -39.20) >DroEre_CAF1 2690 101 - 1 GGAAGGAGCACAUCUUGUGCUGAGCCAGCGUGCUGCAGUGGAGCAU-UCGUGGCAG------------------CUCCAGAUGCAGCUGCUGCAGCACUGAUAAUAAAUUCAUGAUGAAG ..........((((..((((((...(((((.((((((.((((((..-........)------------------)))))..)))))))))))))))))(((........))).))))... ( -40.30) >DroYak_CAF1 2333 101 - 1 GGAAGGAGCACAUCUUGUGCUGAGCCAGCGUGCUGCAGUGGUGCAU-UCGUGGCAG------------------CUCCAGAUGCAGCUGCUGCAGCACUGAUAAUAAAUUCAUGAUGAAG ((....(((((.....)))))...))...(((((((((..((((((-(..(((...------------------..))))))))).)..)))))))))(((........)))........ ( -39.20) >DroAna_CAF1 2290 120 - 1 GCAAGGACCUCAUCUUGUGCUGAGCCAGCGUGCUGCCUUGGUGCAUUUCGUGGCAGCAUCAGCAUCAGCUUCACCUCCAGCUGCAUCUGCCCCAGCACUGAUAAUAAAUUCAUGAUGAAG .........(((((..((((((.((..((((((((((.(((......))).))))))))..))..(((((........))))).....))..))))))(((........))).))))).. ( -38.20) >consensus GGAAGGAGCACAUCUUGUGCUGAGCCAGCGUGCUGCAGUGGUGCAU_UCGUGGCAG__________________CUCCAGAUGCAGCUGCUGCAGCACUGAUAAUAAAUUCAUGAUGAAG ..........((((.((.((((...))))(((((((((((((((((....(((.((..................))))).))))).))))))))))))............)).))))... (-30.82 = -31.82 + 1.00)
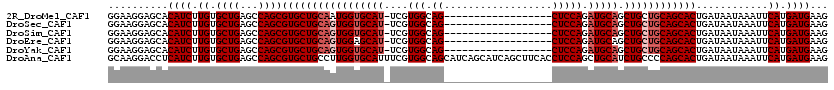
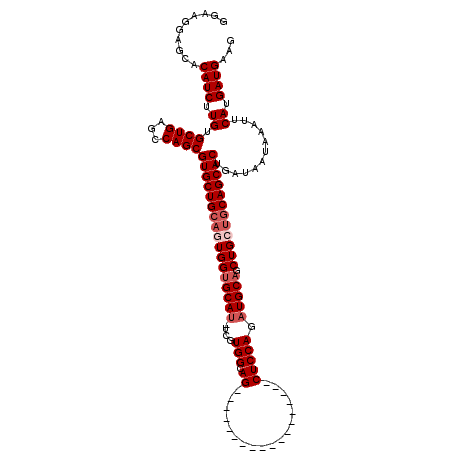
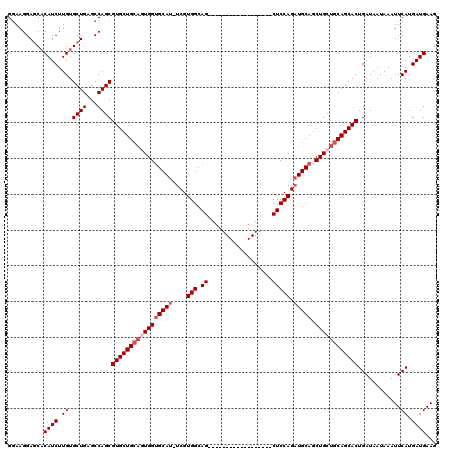
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:25:20 2006