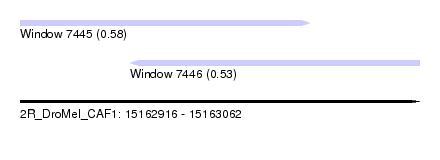
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 15,162,916 – 15,163,062 |
| Length | 146 |
| Max. P | 0.576022 |

| Location | 15,162,916 – 15,163,022 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 85.09 |
| Mean single sequence MFE | -21.74 |
| Consensus MFE | -12.16 |
| Energy contribution | -13.12 |
| Covariance contribution | 0.96 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.576022 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
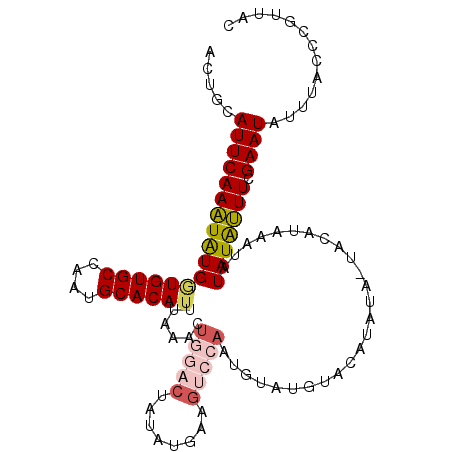
>2R_DroMel_CAF1 15162916 106 + 20766785 ACUGCAUUCAAAUAUGGUGUGCCAAUGCACAUUUAAACUAGACUAUAUGAAGUCCAAUGUAUGUACAUAUA-UACAUAAAUAUAUAUUUCGAAUAUUUACCCGUUAC .....(((((((((((((((((....))))))........((((......))))..((((((((....)))-))))).....))))))).))))............. ( -19.90) >DroSec_CAF1 1327 106 + 1 ACUGCAUUCAAAUAUGGUGUGCCAAUGCACAUUUAAACUGGACGAUAUGAAGUCCAAUGUAUGUACAUAUA-UACAUAAAUAUAUAUUUCGAAUAUUUACCUGUUAC .....(((((((((((((((((....))))))......(((((........)))))((((((((....)))-))))).....))))))).))))............. ( -24.10) >DroSim_CAF1 1323 106 + 1 ACUGCAUUCAAAUAUGGUGUGCCAAUGCACAUUUAAACUGGACUAUAUGAAGUCCAAUGUAUGUACAUAUA-UACAUAAAUAUAUAUUUCGAAUAUUUACCUGUUAC .....(((((((((((((((((....))))))......((((((......))))))((((((((....)))-))))).....))))))).))))............. ( -24.70) >DroEre_CAF1 1683 102 + 1 AAUGCAUUCAAAUGUGCUGUGCCAACGCACAUUUAAGCUGUUCUAUAUGGAGUCCAAUGCAUG----UAUA-UACAUAAAUAUAUGUUUAGAAUAUUUACCCUUUAC ...((.((.((((((((.((....))))))))))))))(((((((..(((...)))..(((((----(((.-.......)))))))).)))))))............ ( -21.10) >DroYak_CAF1 1313 93 + 1 ACUGCAUUCAAGUAUGAUGUGCCAACGCACAUUUAAGCUGGACUAUAUG----------UAUA----UAUAGUACAUGAAUAUAUGCUUAGAAUAUUUACCCUUUAC .....(((((((((((((((((....))))))......((.(((((((.----------...)----)))))).))......))))))).))))............. ( -18.90) >consensus ACUGCAUUCAAAUAUGGUGUGCCAAUGCACAUUUAAACUGGACUAUAUGAAGUCCAAUGUAUGUACAUAUA_UACAUAAAUAUAUAUUUCGAAUAUUUACCCGUUAC .....(((((((((((((((((....))))))......(((((........)))))..........................))))))).))))............. (-12.16 = -13.12 + 0.96)

| Location | 15,162,956 – 15,163,062 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 78.61 |
| Mean single sequence MFE | -24.07 |
| Consensus MFE | -14.55 |
| Energy contribution | -13.75 |
| Covariance contribution | -0.80 |
| Combinations/Pair | 1.39 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.60 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.529398 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
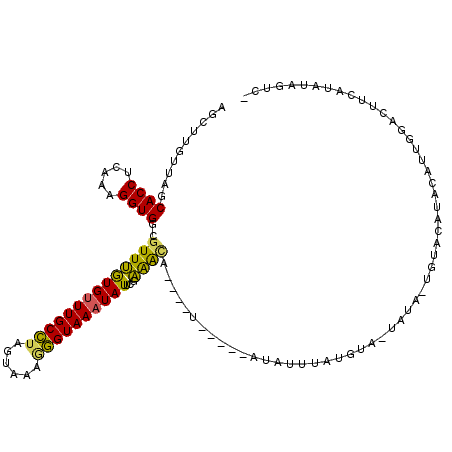
>2R_DroMel_CAF1 15162956 106 - 20766785 AGCUUGUUAGCACCUCAAAGGUGGCGUUUGUGUUUGCCUAGUAACGGGUAAAUAUUCGAAAUA----U-----AUAUUUAUGUA-UAUA-UGUACAUACAUUGGACUUCAUAUAGUC- ..........((((.....)))).((...((((((((((......)))))))))).)).....----.-----.....((((((-(...-.))))))).....((((......))))- ( -22.90) >DroSec_CAF1 1367 106 - 1 AGCUUGUUAGCACCUCAAAGGUGGCGUUUGUGUUUGCCUAGUAACAGGUAAAUAUUCGAAAUA----U-----AUAUUUAUGUA-UAUA-UGUACAUACAUUGGACUUCAUAUCGUC- .(((....)))(((.....)))((((...((((((((((......))))))))))..((((((----(-----((((....)))-))))-)((.((.....)).)))))....))))- ( -21.70) >DroSim_CAF1 1363 106 - 1 AACUUGUUAGCACCUCAAAGGUGGCGUUUGUGUUUGCCUAGUAACAGGUAAAUAUUCGAAAUA----U-----AUAUUUAUGUA-UAUA-UGUACAUACAUUGGACUUCAUAUAGUC- ..........((((.....)))).((...((((((((((......)))))))))).)).....----.-----.....((((((-(...-.))))))).....((((......))))- ( -22.90) >DroEre_CAF1 1723 102 - 1 AGCUUGUUAGCACCUCAAAGGUGGCGUUUGUGUUUGCCUAGUAAAGGGUAAAUAUUCUAAACA----U-----AUAUUUAUGUA-UAUA-----CAUGCAUUGGACUCCAUAUAGAA- .(((....))).........((((.((((((((((((((......)))))))))).....(((----(-----(....))))).-....-----........)))).))))......- ( -21.90) >DroYak_CAF1 1353 93 - 1 AGCUUGUUAGCACCUCAAAGGUGGCGUUUGUGUUUGCCUAGUAAAGGGUAAAUAUUCUAAGCA----U-----AUAUUCAUGUACUAUA-----UAUA----------CAUAUAGUC- .(((((....((((.....))))......((((((((((......))))))))))..))))).----.-----..........((((((-----(...----------.))))))).- ( -23.10) >DroAna_CAF1 1254 117 - 1 GGCUCCUUAGCACCUCAAAGGUGGCAUUCAUGCUUGCUCAGUAAAUAGUAAAUAUUAUGGACAAGUACAUGGAAUAUUUAUGUA-CAAAGUGUAUGUCUUUUGAGCCACAUAAGGGCU (((.(((((....((((((((..((((..((((((.....((..(((((....)))))..))..(((((((((...))))))))-).)))))))))))))))))).....)))))))) ( -31.90) >consensus AGCUUGUUAGCACCUCAAAGGUGGCGUUUGUGUUUGCCUAGUAAAGGGUAAAUAUUCGAAACA____U_____AUAUUUAUGUA_UAUA_UGUACAUACAUUGGACUUCAUAUAGUC_ ..........((((.....))))..((((((((((((((......))))))))))...))))........................................................ (-14.55 = -13.75 + -0.80)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:25:14 2006