| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 15,160,470 – 15,160,570 |
| Length | 100 |
| Max. P | 0.871298 |

| Location | 15,160,470 – 15,160,570 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 74.90 |
| Mean single sequence MFE | -24.67 |
| Consensus MFE | -9.62 |
| Energy contribution | -11.28 |
| Covariance contribution | 1.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.39 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.871298 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
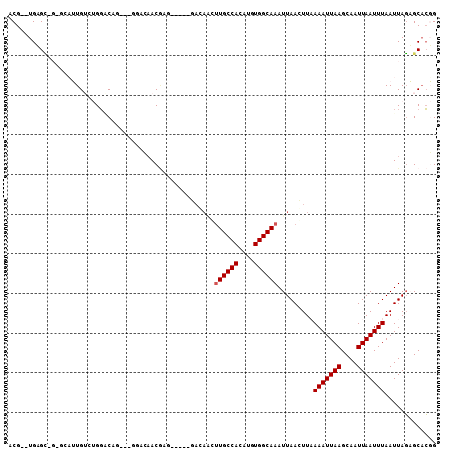
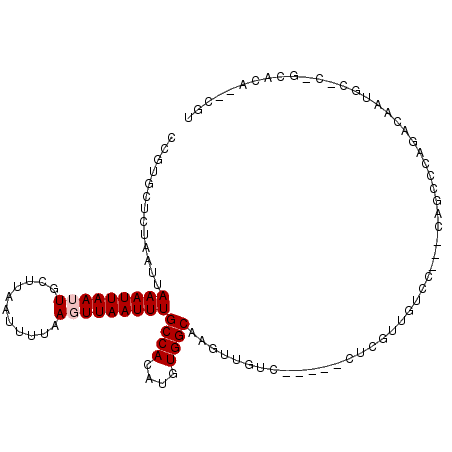
>2R_DroMel_CAF1 15160470 100 + 20766785 ACG--UGUGC-GUGCUUUGUCUGGCCAGAGAGGACAACGAG-----GAGAGGUUGCCACAUGUGGCAAAUUAACUUUAAAUUAAGCAAUUAAUUUAAUUAGAGCACGG ...--....(-((((((((....((((.(..((.((((...-----.....))))))...).)))).........(((((((((....))))))))).))))))))). ( -30.90) >DroVir_CAF1 213556 89 + 1 ACG--UGAGC-----AAUG---GG-CCA---ACAAAACGAG-----CACAACUUGCCACAUGUGGCAAAUUAAGUUAAAAUUAAGCAAUUAAUUUAAUUAGUGCACUG ..(--(..((-----....---.)-)..---)).......(-----(((...((((((....))))))....(((((((.((((....))))))))))).)))).... ( -21.40) >DroYak_CAF1 139556 100 + 1 ACG--UGUGC-GGGAUUUGUGUGGACUGAGAGGACAACGAG-----GAGAAGUUGCCACAUGUGGCAAAUUAACUUUAAAUUAAGCAAUUAAUUUAAUUAGAGCACGG .((--(((.(-..(((((((((((.((....)).((((...-----.....)))))))).....)))))))....(((((((((....)))))))))...).))))). ( -26.00) >DroMoj_CAF1 220633 97 + 1 ACGUGUGAGC-----AAUG---GGGCAA---ACAAAACGAGGCGAACACAACUUGCCACAUGUGGCAAAUUAAGUUAAAAUUAAGCAAUUAAUUUAAUUAGUGCACUG ..((((..((-----..((---......---......))..))..))))...((((((....))))))(((((....(((((((....)))))))..)))))...... ( -20.50) >DroAna_CAF1 136082 90 + 1 ACG--UGAGUAGUGCAUUGUCUGGGCGG--AGGACAAC--------------UUGCCACAUGUGGCAAAUUAACUUAAAAUUAAGCAAUUAAUUUAAUUAGAGCACGG ((.--...)).((((.((((((......--.)))))).--------------((((((....)))))).........(((((((....))))))).......)))).. ( -25.00) >DroPer_CAF1 138974 91 + 1 -CG--UGUGA-GGGCAUUGUCUGCACUC--------ACAAG-----GACAUCAUGCCACAUGUGGCAAAUUAACUUAAAAUUAAGCAAUUAAUUUAAUUAUAGCCGGG -..--(((((-(.(((.....))).)))--------))).(-----(.(....(((((....)))))..........(((((((....))))))).......)))... ( -24.20) >consensus ACG__UGAGC_G_GCAUUGUCUGGACAG___GGACAACGAG_____GACAACUUGCCACAUGUGGCAAAUUAACUUAAAAUUAAGCAAUUAAUUUAAUUAGAGCACGG ....................................................((((((....)))))).........(((((((....)))))))............. ( -9.62 = -11.28 + 1.67)


| Location | 15,160,470 – 15,160,570 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 74.90 |
| Mean single sequence MFE | -20.40 |
| Consensus MFE | -8.60 |
| Energy contribution | -8.93 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.42 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.575991 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
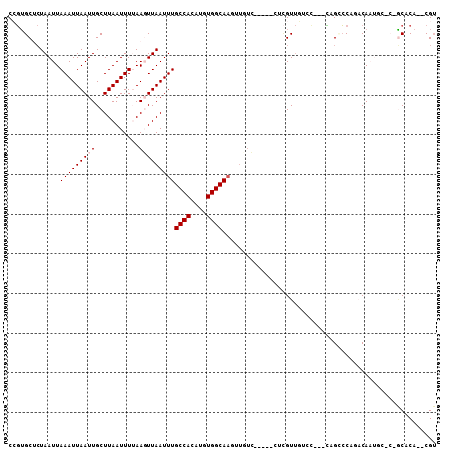
>2R_DroMel_CAF1 15160470 100 - 20766785 CCGUGCUCUAAUUAAAUUAAUUGCUUAAUUUAAAGUUAAUUUGCCACAUGUGGCAACCUCUC-----CUCGUUGUCCUCUCUGGCCAGACAAAGCAC-GCACA--CGU .((((((....(((((((((....))))))))).(((.....((((...(.((((((.....-----...)))).)).)..))))..)))..)))))-)....--... ( -22.90) >DroVir_CAF1 213556 89 - 1 CAGUGCACUAAUUAAAUUAAUUGCUUAAUUUUAACUUAAUUUGCCACAUGUGGCAAGUUGUG-----CUCGUUUUGU---UGG-CC---CAUU-----GCUCA--CGU ..(.((((.....(((((((....)))))))......(((((((((....))))))))))))-----).).....((---.((-(.---....-----))).)--).. ( -20.90) >DroYak_CAF1 139556 100 - 1 CCGUGCUCUAAUUAAAUUAAUUGCUUAAUUUAAAGUUAAUUUGCCACAUGUGGCAACUUCUC-----CUCGUUGUCCUCUCAGUCCACACAAAUCCC-GCACA--CGU ..((((.....(((((((((....))))))))).......((((((....))))))......-----...((((..(.....)..)).)).......-)))).--... ( -18.10) >DroMoj_CAF1 220633 97 - 1 CAGUGCACUAAUUAAAUUAAUUGCUUAAUUUUAACUUAAUUUGCCACAUGUGGCAAGUUGUGUUCGCCUCGUUUUGU---UUGCCC---CAUU-----GCUCACACGU .((.((.......(((((((....))))))).(((.((((((((((....)))))))))).))).)))).....(((---..((..---....-----))..)))... ( -18.30) >DroAna_CAF1 136082 90 - 1 CCGUGCUCUAAUUAAAUUAAUUGCUUAAUUUUAAGUUAAUUUGCCACAUGUGGCAA--------------GUUGUCCU--CCGCCCAGACAAUGCACUACUCA--CGU ..((((..((((((((((((....)))))))..)))))..((((((....))))))--------------((((((..--.......))))))))))......--... ( -19.20) >DroPer_CAF1 138974 91 - 1 CCCGGCUAUAAUUAAAUUAAUUGCUUAAUUUUAAGUUAAUUUGCCACAUGUGGCAUGAUGUC-----CUUGU--------GAGUGCAGACAAUGCCC-UCACA--CG- ...((.(((....(((((((((...........)))))))))((((....))))...))).)-----).(((--------(((.(((.....))).)-)))))--..- ( -23.00) >consensus CCGUGCUCUAAUUAAAUUAAUUGCUUAAUUUUAAGUUAAUUUGCCACAUGUGGCAAGUUGUC_____CUCGUUGUCC___CAGCCCAGACAAUGC_C_GCACA__CGU .............(((((((((...........)))))))))((((....))))...................................................... ( -8.60 = -8.93 + 0.33)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:25:11 2006