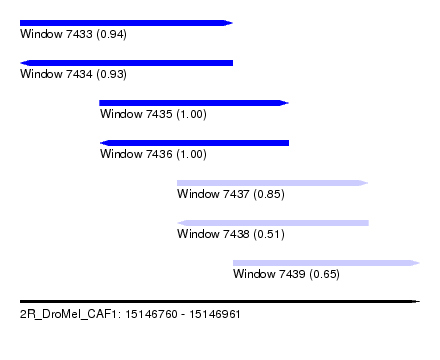
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 15,146,760 – 15,146,961 |
| Length | 201 |
| Max. P | 0.999962 |

| Location | 15,146,760 – 15,146,867 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 78.13 |
| Mean single sequence MFE | -33.08 |
| Consensus MFE | -20.87 |
| Energy contribution | -21.27 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.63 |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.939457 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
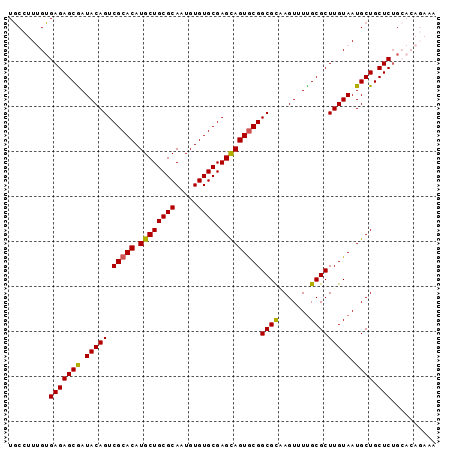
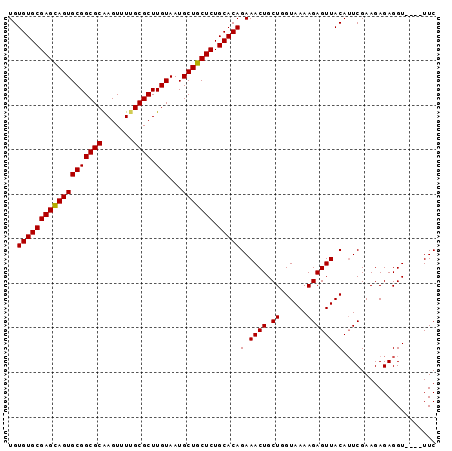
>2R_DroMel_CAF1 15146760 107 + 20766785 ACGUCGUAAUGAGAAAAUGAGAGCUGUAUUAUUUUUGUACUGCCUUUGUGAGAGCGAUACAGUCGCACAUGCUGCGCAAUGUGUGCGAGUAGUGCGGCGCGAGUUUU .((.(((...............(((((((..((((..((.......))..))))..)))))))(((((.((((((((.....)))).))))))))))))))...... ( -33.50) >DroVir_CAF1 183578 81 + 1 AAUUCGUACUGAGAAAAUGAGCGGCGUGCU---------------------CUGCAGUAAAAAACCACAUUUGCCACAUAGUGUGCGAACAGUACAGCAUGU----- .....((((((.......((((.....)))---------------------).................((((((((...))).))))))))))).......----- ( -18.90) >DroSec_CAF1 118970 107 + 1 ACGUCGUAAUGAGAAAAUGAGAGCGGUAUUAUUUUUGUAGUGCCUUUGUGAGAGCGAUACAGUCGCACAUGCUGCGCAAUGUGUGCGAGCAGUGCGGCGCAAGUUUC ......................((((((((((....)))))))).((((....))))....(((((((.((((((((.....)))).)))))))))))))....... ( -37.40) >DroSim_CAF1 104022 107 + 1 ACGUCGUAAUGAGAAAAUGAGAGCGGUAUUAUUUUUGUAGUGCCUUUGUGAGAGCGAUACAGUCGCACAUGCUGCGCAAUGUGUGCGAGCAGUGCGGCGCAAGUUUU ......................((((((((((....)))))))).((((....))))....(((((((.((((((((.....)))).)))))))))))))....... ( -37.40) >DroEre_CAF1 122796 106 + 1 -CGUCGUAAUGAGAAAAUGAGAGCUGUAUUAUUUUUGUAGUGCUUUAGUGAGAGCAAUACAGUCGCACAUGCUGCGCAGUGUGUGCGAGCAGUACGGCGUUAGUUUU -(((((((.((.((((((((........))))))))(((.((((((....)))))).)))..(((((((((((....))))))))))).)).)))))))........ ( -37.40) >DroYak_CAF1 124800 107 + 1 ACGUCGUAAUGAGAAAAUGAGAGCCGUGUUAUUUUUGUAGUACUUUAGUGAGAGCGAUACAGUCGCACAUGCUGCGCAGUGUGUGCGAGCAGUGCGGCGCUAGUUUU .............(((((.((.((((..((....(((((...((((....))))...)))))(((((((((((....)))))))))))..))..)))).)).))))) ( -33.90) >consensus ACGUCGUAAUGAGAAAAUGAGAGCCGUAUUAUUUUUGUAGUGCCUUUGUGAGAGCGAUACAGUCGCACAUGCUGCGCAAUGUGUGCGAGCAGUGCGGCGCAAGUUUU .(((((((.((..............(((((.((((..(.........)..)))).)))))..(((((((((........))))))))).)).)))))))........ (-20.87 = -21.27 + 0.39)


| Location | 15,146,760 – 15,146,867 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 78.13 |
| Mean single sequence MFE | -24.05 |
| Consensus MFE | -14.95 |
| Energy contribution | -16.20 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.928814 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15146760 107 - 20766785 AAAACUCGCGCCGCACUACUCGCACACAUUGCGCAGCAUGUGCGACUGUAUCGCUCUCACAAAGGCAGUACAAAAAUAAUACAGCUCUCAUUUUCUCAUUACGACGU .......((..(((((..((((((.....)))).))...)))))..(((((.(((........))).)))))...........))...................... ( -21.70) >DroVir_CAF1 183578 81 - 1 -----ACAUGCUGUACUGUUCGCACACUAUGUGGCAAAUGUGGUUUUUUACUGCAG---------------------AGCACGCCGCUCAUUUUCUCAGUACGAAUU -----......(((((((...((.(((...)))))...((..((.....))..))(---------------------(((.....)))).......))))))).... ( -21.10) >DroSec_CAF1 118970 107 - 1 GAAACUUGCGCCGCACUGCUCGCACACAUUGCGCAGCAUGUGCGACUGUAUCGCUCUCACAAAGGCACUACAAAAAUAAUACCGCUCUCAUUUUCUCAUUACGACGU ((((...(((.(((((((((((((.....)))).)))).)))))..((((..(((........)))..))))..........)))......))))............ ( -25.90) >DroSim_CAF1 104022 107 - 1 AAAACUUGCGCCGCACUGCUCGCACACAUUGCGCAGCAUGUGCGACUGUAUCGCUCUCACAAAGGCACUACAAAAAUAAUACCGCUCUCAUUUUCUCAUUACGACGU .......(((.(((((((((((((.....)))).)))).)))))..((((..(((........)))..))))..........)))...................... ( -25.60) >DroEre_CAF1 122796 106 - 1 AAAACUAACGCCGUACUGCUCGCACACACUGCGCAGCAUGUGCGACUGUAUUGCUCUCACUAAAGCACUACAAAAAUAAUACAGCUCUCAUUUUCUCAUUACGACG- .........(((((((((((((((.....)))).)))).)))))..((((.((((........)))).))))...........)).....................- ( -24.30) >DroYak_CAF1 124800 107 - 1 AAAACUAGCGCCGCACUGCUCGCACACACUGCGCAGCAUGUGCGACUGUAUCGCUCUCACUAAAGUACUACAAAAAUAACACGGCUCUCAUUUUCUCAUUACGACGU ((((..((.(((((((((((((((.....)))).)))).))))...((((..(((........)))..))))..........))).))..))))............. ( -25.70) >consensus AAAACUAGCGCCGCACUGCUCGCACACAUUGCGCAGCAUGUGCGACUGUAUCGCUCUCACAAAGGCACUACAAAAAUAAUACAGCUCUCAUUUUCUCAUUACGACGU .........(((((((((((((((.....)))).)))).)))))..((((..(((........)))..))))...........))...................... (-14.95 = -16.20 + 1.25)


| Location | 15,146,800 – 15,146,895 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 88.75 |
| Mean single sequence MFE | -39.98 |
| Consensus MFE | -34.94 |
| Energy contribution | -34.66 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.10 |
| Mean z-score | -3.77 |
| Structure conservation index | 0.87 |
| SVM decision value | 4.42 |
| SVM RNA-class probability | 0.999894 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
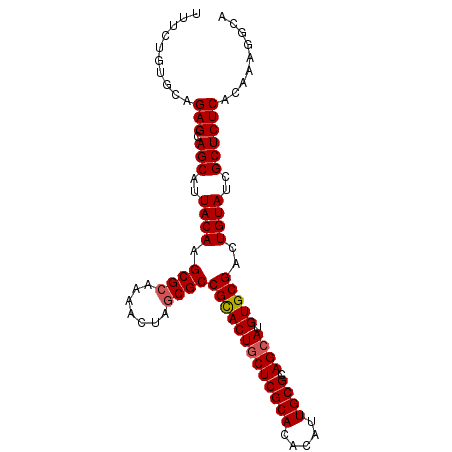
>2R_DroMel_CAF1 15146800 95 + 20766785 UGCCUUUGUGAGAGCGAUACAGUCGCACAUGCUGCGCAAUGUGUGCGAGUAGUGCGGCGCGAGUUUUGCGCUUGUAAUGCUGCUCUGCACAGAAA ....((((((((((((.....(((((((.((((((((.....)))).)))))))))))((((((.....)))))).....)))))).)))))).. ( -41.00) >DroSec_CAF1 119010 95 + 1 UGCCUUUGUGAGAGCGAUACAGUCGCACAUGCUGCGCAAUGUGUGCGAGCAGUGCGGCGCAAGUUUCGCGCUUGUAAUGCUGCUCUGCACAGAAA ....((((((((((((.....(((((((.((((((((.....)))).)))))))))))((((((.....)))))).....)))))).)))))).. ( -43.30) >DroSim_CAF1 104062 95 + 1 UGCCUUUGUGAGAGCGAUACAGUCGCACAUGCUGCGCAAUGUGUGCGAGCAGUGCGGCGCAAGUUUUGCGCUUGUAAUGCUGCUCUGCACAGAAA ....((((((((((((.....(((((((.((((((((.....)))).)))))))))))((((((.....)))))).....)))))).)))))).. ( -43.30) >DroEre_CAF1 122835 87 + 1 UGCUUUAGUGAGAGCAAUACAGUCGCACAUGCUGCGCAGUGUGUGCGAGCAGUACGGCGUUAGUUUUGCGCUUGUAAUGCUGCUCUG-------- ..........((((((......(((((((((((....)))))))))))(((.((((((((.......)))).)))).))))))))).-------- ( -36.50) >DroYak_CAF1 124840 85 + 1 UACUUUAGUGAGAGCGAUACAGUCGCACAUGCUGCGCAGUGUGUGCGAGCAGUGCGGCGCUAGUUUUGCGCUUGUAAUGCUGCUC---------- .........(((((((.(((((.(((((.((((((((.....)))).)))))))))((((.......))))))))).)))).)))---------- ( -35.80) >consensus UGCCUUUGUGAGAGCGAUACAGUCGCACAUGCUGCGCAAUGUGUGCGAGCAGUGCGGCGCAAGUUUUGCGCUUGUAAUGCUGCUCUGCACAGAAA .........(((((((.(((((.(((((.((((((((.....)))).)))))))))((((.......))))))))).)))).))).......... (-34.94 = -34.66 + -0.28)

| Location | 15,146,800 – 15,146,895 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 88.75 |
| Mean single sequence MFE | -34.88 |
| Consensus MFE | -31.98 |
| Energy contribution | -32.22 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -4.11 |
| Structure conservation index | 0.92 |
| SVM decision value | 4.93 |
| SVM RNA-class probability | 0.999962 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
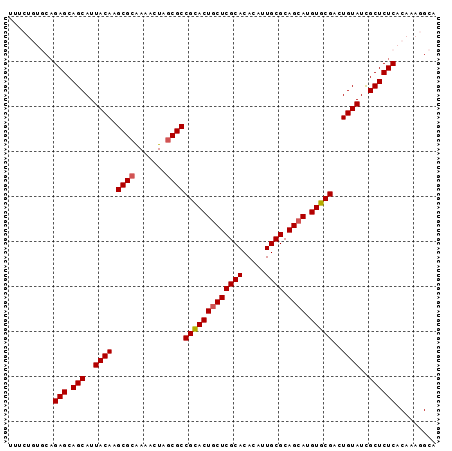
>2R_DroMel_CAF1 15146800 95 - 20766785 UUUCUGUGCAGAGCAGCAUUACAAGCGCAAAACUCGCGCCGCACUACUCGCACACAUUGCGCAGCAUGUGCGACUGUAUCGCUCUCACAAAGGCA ....((((.(((((.....((((.((((.......))))(((((..((((((.....)))).))...)))))..))))..)))))))))...... ( -32.00) >DroSec_CAF1 119010 95 - 1 UUUCUGUGCAGAGCAGCAUUACAAGCGCGAAACUUGCGCCGCACUGCUCGCACACAUUGCGCAGCAUGUGCGACUGUAUCGCUCUCACAAAGGCA ....((((.(((((.....((((.((((((...))))))(((((((((((((.....)))).)))).)))))..))))..)))))))))...... ( -38.30) >DroSim_CAF1 104062 95 - 1 UUUCUGUGCAGAGCAGCAUUACAAGCGCAAAACUUGCGCCGCACUGCUCGCACACAUUGCGCAGCAUGUGCGACUGUAUCGCUCUCACAAAGGCA ....((((.(((((.....((((.(((((.....)))))(((((((((((((.....)))).)))).)))))..))))..)))))))))...... ( -38.60) >DroEre_CAF1 122835 87 - 1 --------CAGAGCAGCAUUACAAGCGCAAAACUAACGCCGUACUGCUCGCACACACUGCGCAGCAUGUGCGACUGUAUUGCUCUCACUAAAGCA --------..(((.((((.((((.(((.........)))(((((((((((((.....)))).)))).)))))..)))).)))))))......... ( -30.80) >DroYak_CAF1 124840 85 - 1 ----------GAGCAGCAUUACAAGCGCAAAACUAGCGCCGCACUGCUCGCACACACUGCGCAGCAUGUGCGACUGUAUCGCUCUCACUAAAGUA ----------(((.(((..((((.((((.......))))(((((((((((((.....)))).)))).)))))..))))..))))))......... ( -34.70) >consensus UUUCUGUGCAGAGCAGCAUUACAAGCGCAAAACUAGCGCCGCACUGCUCGCACACAUUGCGCAGCAUGUGCGACUGUAUCGCUCUCACAAAGGCA ..........(((.(((..((((.((((.......))))(((((((((((((.....)))).)))).)))))..))))..))))))......... (-31.98 = -32.22 + 0.24)

| Location | 15,146,839 – 15,146,935 |
|---|---|
| Length | 96 |
| Sequences | 3 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 93.66 |
| Mean single sequence MFE | -32.27 |
| Consensus MFE | -28.15 |
| Energy contribution | -27.93 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.846316 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
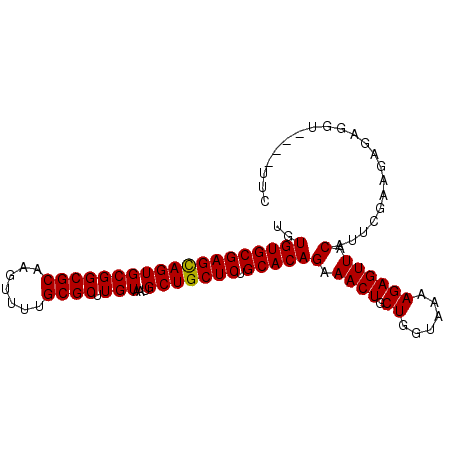
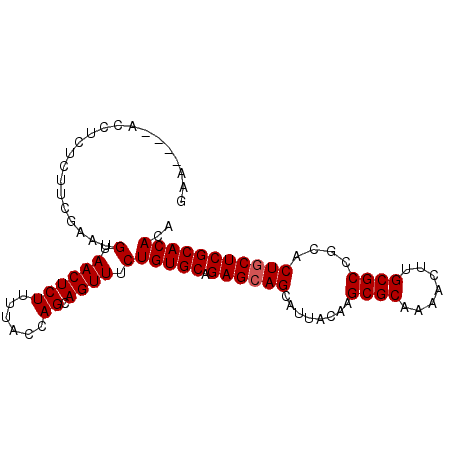
>2R_DroMel_CAF1 15146839 96 + 20766785 UGUGUGCGAGUAGUGCGGCGCGAGUUUUGCGCUUGUAAUGCUGCUCUGCACAGAAACUGCUGGUAAAAGAGUUACAAUCGAAGAGAGGUUUGUUUC ..(((((((((((((((((((((...)))))).)))...))))))).)))))(((((.(((.((((.....))))..((.....)))))..))))) ( -30.70) >DroSec_CAF1 119049 92 + 1 UGUGUGCGAGCAGUGCGGCGCAAGUUUCGCGCUUGUAAUGCUGCUCUGCACAGAAACUGCUUGUAAAAGAGUUACAUUCGAAGAGAGGU----UUC ..(((((((((((((((((((.......)))).)))...))))))).)))))((((((.(((......((((...))))...))).)))----))) ( -33.10) >DroSim_CAF1 104101 92 + 1 UGUGUGCGAGCAGUGCGGCGCAAGUUUUGCGCUUGUAAUGCUGCUCUGCACAGAAACUGCUGGUAAAAGAGUUACAUUCGAAGAGAGGU----UUC ..((((((((((((((((((((.....))))).)))...))))))).)))))((((((.((.((((.....))))......))...)))----))) ( -33.00) >consensus UGUGUGCGAGCAGUGCGGCGCAAGUUUUGCGCUUGUAAUGCUGCUCUGCACAGAAACUGCUGGUAAAAGAGUUACAUUCGAAGAGAGGU____UUC ..(((((((((((((((((((.......)))).)))...))))))).)))))(.((((.((......)))))).)..................... (-28.15 = -27.93 + -0.22)

| Location | 15,146,839 – 15,146,935 |
|---|---|
| Length | 96 |
| Sequences | 3 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 93.66 |
| Mean single sequence MFE | -22.47 |
| Consensus MFE | -19.27 |
| Energy contribution | -19.60 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.86 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.511148 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
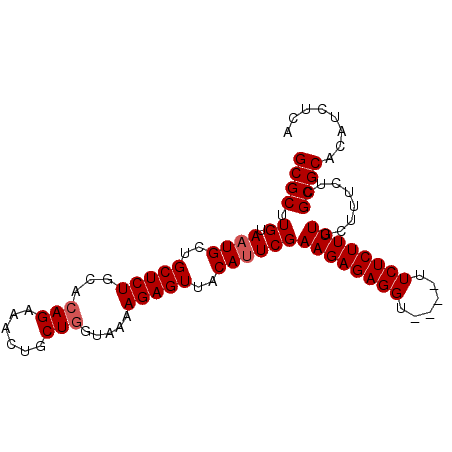
>2R_DroMel_CAF1 15146839 96 - 20766785 GAAACAAACCUCUCUUCGAUUGUAACUCUUUUACCAGCAGUUUCUGUGCAGAGCAGCAUUACAAGCGCAAAACUCGCGCCGCACUACUCGCACACA (((((.....((.....))..((((.....)))).....)))))(((((.(((.((........((((.......))))....)).)))))))).. ( -17.20) >DroSec_CAF1 119049 92 - 1 GAA----ACCUCUCUUCGAAUGUAACUCUUUUACAAGCAGUUUCUGUGCAGAGCAGCAUUACAAGCGCGAAACUUGCGCCGCACUGCUCGCACACA (((----((.((.....)).(((((.....)))))....)))))(((((.((((((........((((((...))))))....))))))))))).. ( -25.90) >DroSim_CAF1 104101 92 - 1 GAA----ACCUCUCUUCGAAUGUAACUCUUUUACCAGCAGUUUCUGUGCAGAGCAGCAUUACAAGCGCAAAACUUGCGCCGCACUGCUCGCACACA (((----((.((.....))..((((.....)))).....)))))(((((.((((((........(((((.....)))))....))))))))))).. ( -24.30) >consensus GAA____ACCUCUCUUCGAAUGUAACUCUUUUACCAGCAGUUUCUGUGCAGAGCAGCAUUACAAGCGCAAAACUUGCGCCGCACUGCUCGCACACA .....................(.((((((......)).)))).)(((((.((((((........((((.......))))....))))))))))).. (-19.27 = -19.60 + 0.33)


| Location | 15,146,867 – 15,146,961 |
|---|---|
| Length | 94 |
| Sequences | 3 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 95.68 |
| Mean single sequence MFE | -25.20 |
| Consensus MFE | -23.67 |
| Energy contribution | -24.33 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.652785 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15146867 94 + 20766785 GCGCUUGUAAUGCUGCUCUGCACAGAAACUGCUGGUAAAAGAGUUACAAUCGAAGAGAGGUUUGUUUCUCUUUUGCUUUCUCGCGCACAUCUCA ((((......(((......)))(((......)))(((((((((..((((((.......)).))))..)))))))))......))))........ ( -26.00) >DroSec_CAF1 119077 90 + 1 GCGCUUGUAAUGCUGCUCUGCACAGAAACUGCUUGUAAAAGAGUUACAUUCGAAGAGAGGU----UUCUCUUUUGCUUUCUCGCGCACAUCUCA ((((......(((......))).(((((......(((((((((..((.(((.....)))))----..)))))))))))))).))))........ ( -24.10) >DroSim_CAF1 104129 90 + 1 GCGCUUGUAAUGCUGCUCUGCACAGAAACUGCUGGUAAAAGAGUUACAUUCGAAGAGAGGU----UUCUCUUUUGCUUUCUCGCGCACAUCUCA ((((......(((......)))(((......)))(((((((((..((.(((.....)))))----..)))))))))......))))........ ( -25.50) >consensus GCGCUUGUAAUGCUGCUCUGCACAGAAACUGCUGGUAAAAGAGUUACAUUCGAAGAGAGGU____UUCUCUUUUGCUUUCUCGCGCACAUCUCA ((((.((.((((..(((((...(((......))).....)))))..))))))((((((((......))))))))........))))........ (-23.67 = -24.33 + 0.67)

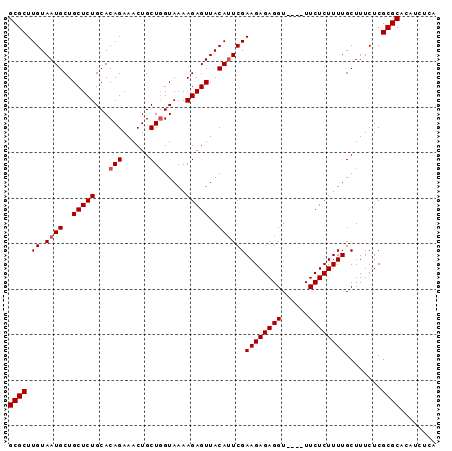
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:25:08 2006