| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 15,104,039 – 15,104,134 |
| Length | 95 |
| Max. P | 0.966129 |

| Location | 15,104,039 – 15,104,134 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 68.78 |
| Mean single sequence MFE | -30.18 |
| Consensus MFE | -12.90 |
| Energy contribution | -12.48 |
| Covariance contribution | -0.42 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.43 |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.923149 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
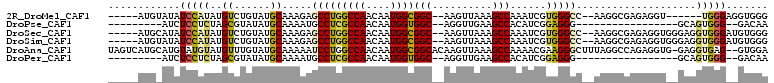
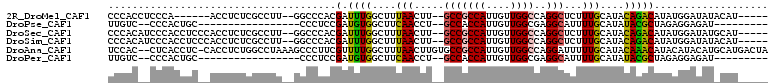
>2R_DroMel_CAF1 15104039 95 + 20766785 -----AUGUAUAUCCAUAUGUCUGUAUGCAAAGAGCCUGGCCAACAAUGGCGGC--AAGUUAAAGCCAAAUCGUGGGCC--AAGGCGAGAGGU------UGGGAGGUGGG -----........((((...(((....((.....((((((((.((.((...(((--........)))..)).)).))))--.)))).....))------.)))..)))). ( -27.40) >DroPse_CAF1 85744 80 + 1 ---------AUCUCCUCUAGCGUAUAUGCAAAAUGCCUCGCCAACAAUGGUGGC--AGGUUGAAGCCACAUCGGAGGG-----------------GCAGUGGG--GACAA ---------.(((((.((.(((....))).....(((((.((.......(((((--........)))))...)).)))-----------------)))).)))--))... ( -28.20) >DroSec_CAF1 80654 101 + 1 -----AUGCAUAUCCAUAUGUCUGUAUGCAAAGAGCCUGGCCAACAAUGGCGGC--AAGUUAAAGCCAAAUCGUGGGCC--AAGGCGAGAGGUGGGAGGUGGGAUGUGGG -----...(((((((.(((.(((...(((.....((((((((.((.((...(((--........)))..)).)).))))--.)))).....)))))).)))))))))).. ( -35.00) >DroSim_CAF1 66533 101 + 1 -----AUGUAUAUCCAUAUGUCUGUAUGCAAAGAGCCUGGCCAACAAUGGCGGC--AAGUUAAAGCCAAAUCGUGGGCC--AAGGCGAGAGGUGGGAGGUGGGAUGUGGG -----...(((((((.(((.(((...(((.....((((((((.((.((...(((--........)))..)).)).))))--.)))).....)))))).)))))))))).. ( -32.90) >DroAna_CAF1 84074 107 + 1 UAGUCAUGCAUGUAUGUAUGUUUGUAUGCAAAAAUCCUGGCCAACAAUGGCGGCACAAGUUAAAGCCAAAACGAAGGGCUUUAGGCCAGAGGUG-GAGGUGAG--GUGGA ......((((((((........)))))))).....(((.(((..((.((((.((....))(((((((.........))))))).))))....))-..))).))--).... ( -29.40) >DroPer_CAF1 85227 80 + 1 ---------AUCUCCUCUAGCGUAUAUGCAAAAUGCCUCGCCAACAAUGGUGGC--AGGUUGAAGCCACAUCGGAGGG-----------------GCAGUGGG--GACAA ---------.(((((.((.(((....))).....(((((.((.......(((((--........)))))...)).)))-----------------)))).)))--))... ( -28.20) >consensus _____AUG_AUAUCCAUAUGUCUGUAUGCAAAAAGCCUGGCCAACAAUGGCGGC__AAGUUAAAGCCAAAUCGUAGGCC__AAGGCGAGAGGU__GAGGUGGG__GUGGA ............(((((..((......)).....(((((((((....))))(((..........)))......)))))....................)))))....... (-12.90 = -12.48 + -0.42)
| Location | 15,104,039 – 15,104,134 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 68.78 |
| Mean single sequence MFE | -25.47 |
| Consensus MFE | -7.88 |
| Energy contribution | -7.18 |
| Covariance contribution | -0.69 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.31 |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.966129 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
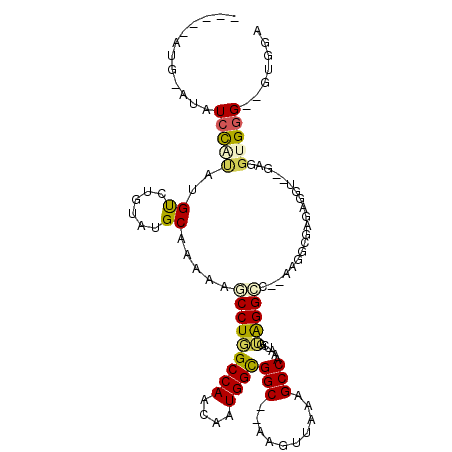
>2R_DroMel_CAF1 15104039 95 - 20766785 CCCACCUCCCA------ACCUCUCGCCUU--GGCCCACGAUUUGGCUUUAACUU--GCCGCCAUUGUUGGCCAGGCUCUUUGCAUACAGACAUAUGGAUAUACAU----- ........(((------.......((((.--((((.(((((.((((........--))))..))))).))))))))..((((....))))....)))........----- ( -26.70) >DroPse_CAF1 85744 80 - 1 UUGUC--CCCACUGC-----------------CCCUCCGAUGUGGCUUCAACCU--GCCACCAUUGUUGGCGAGGCAUUUUGCAUAUACGCUAGAGGAGAU--------- ..(((--.((..(((-----------------(.(.((((((((((........--)))))....))))).).))))(((.((......)).))))).)))--------- ( -24.10) >DroSec_CAF1 80654 101 - 1 CCCACAUCCCACCUCCCACCUCUCGCCUU--GGCCCACGAUUUGGCUUUAACUU--GCCGCCAUUGUUGGCCAGGCUCUUUGCAUACAGACAUAUGGAUAUGCAU----- ........................((((.--((((.(((((.((((........--))))..))))).))))))))....((((((((......))..)))))).----- ( -29.10) >DroSim_CAF1 66533 101 - 1 CCCACAUCCCACCUCCCACCUCUCGCCUU--GGCCCACGAUUUGGCUUUAACUU--GCCGCCAUUGUUGGCCAGGCUCUUUGCAUACAGACAUAUGGAUAUACAU----- .....((((...............((((.--((((.(((((.((((........--))))..))))).))))))))..((((....)))).....))))......----- ( -27.40) >DroAna_CAF1 84074 107 - 1 UCCAC--CUCACCUC-CACCUCUGGCCUAAAGCCCUUCGUUUUGGCUUUAACUUGUGCCGCCAUUGUUGGCCAGGAUUUUUGCAUACAAACAUACAUACAUGCAUGACUA .....--.(((..((-(......(((((((((((.........)))))))....).)))((((....))))..)))....(((((..............))))))))... ( -21.44) >DroPer_CAF1 85227 80 - 1 UUGUC--CCCACUGC-----------------CCCUCCGAUGUGGCUUCAACCU--GCCACCAUUGUUGGCGAGGCAUUUUGCAUAUACGCUAGAGGAGAU--------- ..(((--.((..(((-----------------(.(.((((((((((........--)))))....))))).).))))(((.((......)).))))).)))--------- ( -24.10) >consensus CCCAC__CCCACCUC__ACCUCUCGCCUU__GCCCCACGAUUUGGCUUUAACUU__GCCGCCAUUGUUGGCCAGGCUCUUUGCAUACAGACAUAUGGAUAU_CAU_____ ......................................(.((((....(((.....(((((((....))))..)))...)))....)))))................... ( -7.88 = -7.18 + -0.69)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:24:50 2006