
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 15,093,723 – 15,093,840 |
| Length | 117 |
| Max. P | 0.969431 |

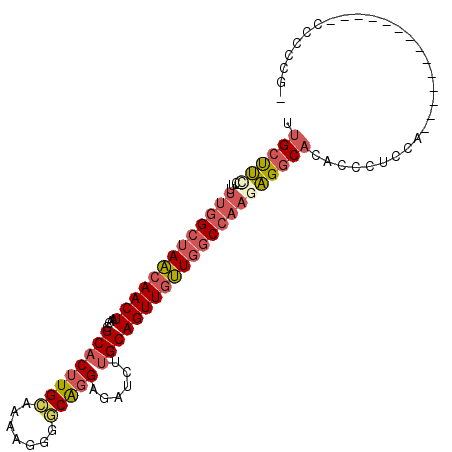
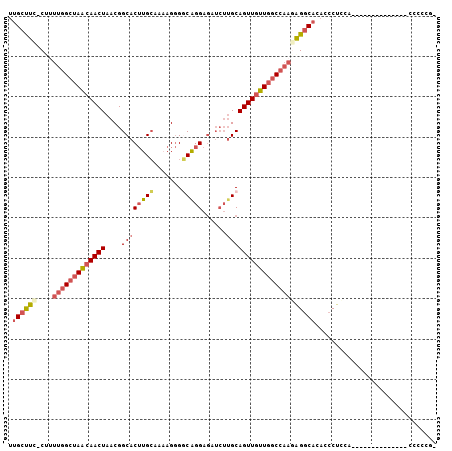
| Location | 15,093,723 – 15,093,813 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 76.82 |
| Mean single sequence MFE | -30.62 |
| Consensus MFE | -22.89 |
| Energy contribution | -23.92 |
| Covariance contribution | 1.03 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.64 |
| SVM RNA-class probability | 0.969431 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15093723 90 - 20766785 UUGCUUC-CUUUUGGCUAACAACUAACGGCACUUGCAGAAGGGACGGGAGAUCUUGCAGUUGUUGGCCAAGAGGCACACCCUCCU--------------CCCCCG- ......(-((((((((((((((((....((....))....(((((....).))))..)))))))))))))))))...........--------------......- ( -30.20) >DroSec_CAF1 70310 89 - 1 UUGCUUC-CU-UUGGCUAACAACUAACGGCACUUGCAAAAGGGGCAGGAGAUCUAGCAGUUGUUGGCCAAGAGGCACACCCUCCG--------------CCCCCG- .((((((-..-(((((((((((((...(((.(((((.......))))).).))....))))))))))))))))))).........--------------......- ( -30.80) >DroSim_CAF1 48257 90 - 1 UUGCUUC-CUUUUGGCUAACAACUAACGGCACUUGCAAAAGGGGCAGGAGAUCUUGCAGUUGUUGGCCAAGAGGCACACCCUCCG--------------CCCCCG- ......(-((((((((((((((((....((....)).......(((((....))))))))))))))))))))))...........--------------......- ( -34.60) >DroEre_CAF1 70684 90 - 1 UUGCUUU-CGUUUGGCUAACAACUAACGGCACUUGCAACAGGGGCAGGAGAUCUUGCAGUUGUUGGCCAACAGGCACACCCUCCU--------------CCCCCG- .(((((.-...(((((((((((((....((....)).......(((((....)))))))))))))))))).))))).........--------------......- ( -30.50) >DroYak_CAF1 72926 97 - 1 UUGCUUU-CGUUUGGCUAACAACUAACGGCACUUGCCAAAGGGGCAGGAGAUCUUGCAGUUGUUGGCCAAGAGGCACACCCUUCACCUC---UUC----CCCCCG- .((((((-...(((((((((((((...(((....)))......(((((....)))))))))))))))))))))))).............---...----......- ( -35.90) >DroPer_CAF1 74051 102 - 1 UUGCCCCGCUUUGGGCCAGCAACUAACGGCACAUGGUG-AGAGCCAGGCCAAAAUGCAGUUACUCAC---UGGUCUCUACCUCUACCUCCCUUUCCCCUCUCUCAC ..((((......))))...........(((...((((.-...)))).)))......((((.....))---)).................................. ( -21.70) >consensus UUGCUUC_CUUUUGGCUAACAACUAACGGCACUUGCAAAAGGGGCAGGAGAUCUUGCAGUUGUUGGCCAAGAGGCACACCCUCCA______________CCCCCG_ .((((((....(((((((((((((....((((((((.......)))))......)))))))))))))))))))))).............................. (-22.89 = -23.92 + 1.03)

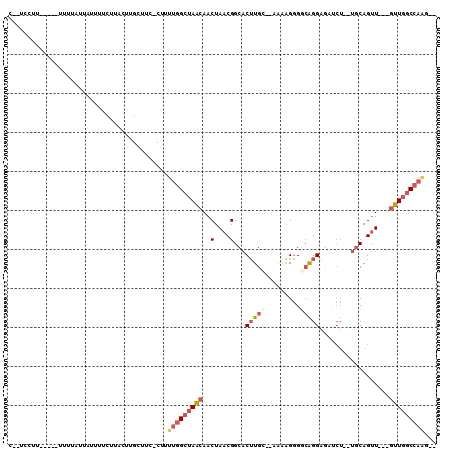
| Location | 15,093,743 – 15,093,840 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 70.90 |
| Mean single sequence MFE | -26.30 |
| Consensus MFE | -10.46 |
| Energy contribution | -11.35 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.40 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.649125 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15093743 97 - 20766785 C--UCCUUUUUU-UUUUAUUAUUUUCUUACUUGCUUC-CUUUUGGCUAACAACUAACGGCACUUGC--AGAAGGGACGGGAGAUCU--UGCAGUU---GUUGGCCAAG-- .--.........-........................-...(((((((((((((....((....))--....(((((....).)))--)..))))---))))))))).-- ( -22.90) >DroSec_CAF1 70330 92 - 1 C--UUCUU-----UUUUAUUAUUUUCUUUCUUGCUUC-CU-UUGGCUAACAACUAACGGCACUUGC--AAAAGGGGCAGGAGAUCU--AGCAGUU---GUUGGCCAAG-- .--.....-----........................-..-(((((((((((((...(((.(((((--.......))))).).)).--...))))---))))))))).-- ( -23.80) >DroSim_CAF1 48277 93 - 1 C--UUCUU-----UUUUAUUAUUUUCUUACUUGCUUC-CUUUUGGCUAACAACUAACGGCACUUGC--AAAAGGGGCAGGAGAUCU--UGCAGUU---GUUGGCCAAG-- .--.....-----........................-...(((((((((((((....((....))--.......(((((....))--)))))))---))))))))).-- ( -27.30) >DroEre_CAF1 70704 95 - 1 C--UCCUUUUU---UUUAUUAUUUUCUUACUUGCUUU-CGUUUGGCUAACAACUAACGGCACUUGC--AACAGGGGCAGGAGAUCU--UGCAGUU---GUUGGCCAAC-- .--........---.......................-...(((((((((((((....((....))--.......(((((....))--)))))))---))))))))).-- ( -27.00) >DroAna_CAF1 72324 104 - 1 CGCUCCUUUAUUUUUGUAUAAUUUUUU-UUUCG-----CUUUUGGCUAACAACUAACGGCACUUGGGGAGAGGGGGAGGGACAACUGCUACAGAUGCAGUUGGCCAGGCC ..(((((((((......))))....((-((((.-----((....(((..........)))....)).)))))))))))((.(((((((.......))))))).))..... ( -27.10) >DroPer_CAF1 74086 86 - 1 C--CGAUU-----CUGC----CUCGCUGCCUUGCCCCGCUUUGGGCCAGCAACUAACGGCACAUGG--UG-AGAGCCAGGCCAAAA--UGCAGUU---ACUCAC---U-- .--.((..-----((((----...((((....((((......)))))))).......(((...(((--(.-...)))).)))....--.))))..---..))..---.-- ( -29.70) >consensus C__UCCUU_____UUUUAUUAUUUUCUUACUUGCUUC_CUUUUGGCUAACAACUAACGGCACUUGC__AAAAGGGGCAGGAGAUCU__UGCAGUU___GUUGGCCAAG__ .........................................(((((((((..(....)...(((((.........)))))..................)))))))))... (-10.46 = -11.35 + 0.89)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:24:47 2006