
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 15,079,242 – 15,079,334 |
| Length | 92 |
| Max. P | 0.863213 |

| Location | 15,079,242 – 15,079,334 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.70 |
| Mean single sequence MFE | -24.57 |
| Consensus MFE | -21.63 |
| Energy contribution | -21.47 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.863213 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
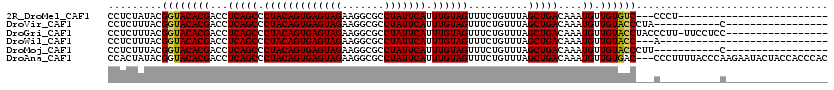
>2R_DroMel_CAF1 15079242 92 - 20766785 CCUCUAUACGGUACACGACCUCAGCCCUACAGUGAGUAGAAGGCGCCUAUUCAUUUGUAGUUUCUGUUUAGCUGACAAAUGUUGUGUC---CCCU------------------------- .........((.(((((((.(((((.(((((((((((((.......))))))).))))))..........))))).....))))))).---))..------------------------- ( -25.80) >DroVir_CAF1 83640 92 - 1 CCUCUUUACGGUACACGACCUCAGCCCUACAGUGAGUAGAAGGCGCCUAUUCAUUUGUAGUUUCUGUUUAGCUGACAAAUGUUGUACCCUA-----------C----------------- .........((((((((...(((((.(((((((((((((.......))))))).))))))..........)))))....)).))))))...-----------.----------------- ( -23.70) >DroGri_CAF1 78475 102 - 1 CCUCUUUACGGUACACGACCUCAGCCCUACAGUGAGUAGAAGGCGCCUAUUCAUUUGUAGUUUCUGUUUAGCUGACAAAUGUUGUACCUACCCUU-UUCCUCC----------------- .........((((((((...(((((.(((((((((((((.......))))))).))))))..........)))))....)).)))))).......-.......----------------- ( -24.10) >DroWil_CAF1 107046 89 - 1 CCUCUUUACGGUACACGACCUCAGCCCUACAGUGAGUAGAAGGCGCCUAUUCAUUUGUAGUUUCUGUUUAGCUGACAAAUGUUGUACC---A---------------------------- .........((((((((...(((((.(((((((((((((.......))))))).))))))..........)))))....)).))))))---.---------------------------- ( -24.60) >DroMoj_CAF1 93894 92 - 1 CCUCUUUACGGUACACGACCUCAGCCCUACAGUGAGUAGAAGGCGCCUAUUCAUUUGUAGUUUCUGUUUAGCUGACAAAUGUUGUACCCUU-----------C----------------- .........((((((((...(((((.(((((((((((((.......))))))).))))))..........)))))....)).))))))...-----------.----------------- ( -23.70) >DroAna_CAF1 58046 117 - 1 CCACUAUACGGUACACGACCUCAGCCCUACAGUGAGUAGAAGGCGCCUAUUCAUUUGUAGUUUCUGUUUAGCUGACAAAUGUUGUGAC---CCCUUUUACCCAAGAAUACUACCACCCAC .........(((.((((((.(((((.(((((((((((((.......))))))).))))))..........))))).....))))))))---)............................ ( -25.50) >consensus CCUCUUUACGGUACACGACCUCAGCCCUACAGUGAGUAGAAGGCGCCUAUUCAUUUGUAGUUUCUGUUUAGCUGACAAAUGUUGUACC___C__________C_________________ .........((((((((...(((((.(((((((((((((.......))))))).))))))..........)))))....)).))))))................................ (-21.63 = -21.47 + -0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:24:43 2006