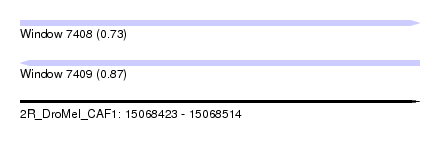
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 15,068,423 – 15,068,514 |
| Length | 91 |
| Max. P | 0.872311 |

| Location | 15,068,423 – 15,068,514 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 80.99 |
| Mean single sequence MFE | -31.95 |
| Consensus MFE | -18.44 |
| Energy contribution | -18.67 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.725161 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
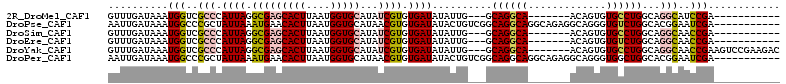
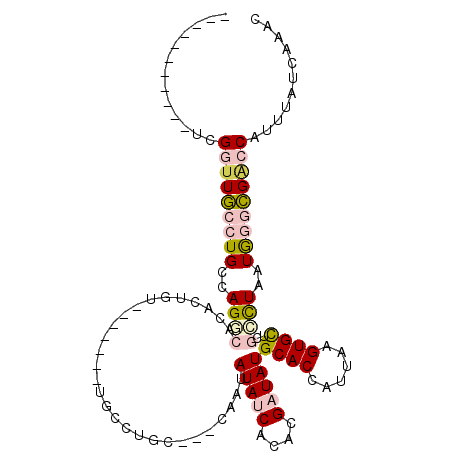
>2R_DroMel_CAF1 15068423 91 + 20766785 GUUUGAUAAAUGGUCGCCCAUUAGGCGAGCACUUAAUGGUGCAUAUCGUGUGAUAUAUUG---GCAGGCA-------ACAGUGUGCCUGGCAGGCAUCCGA----------- ..........(((..(((.((((.(((((((((....)))))...)))).))))....((---.((((((-------......)))))).)))))..))).----------- ( -33.90) >DroPse_CAF1 48874 101 + 1 AAUUGAUAAAUGGCCCGCUAUUAAAUGAACACUUAAUGGUGCAUAACGUGUGAUAUACUGUCGGCAGGCAGGCAGAGGCAGGGUGUCUGGCACGGAAUCGA----------- ..(((((........(((((((((........))))))))).....(((((((((..(((((.((......))...)))))..))))..)))))..)))))----------- ( -28.80) >DroSim_CAF1 23136 91 + 1 GUUUGAUAAAUGGUCGCCCAUUAGGCGAGCACUUAAUGGUGCAUAUCGUGUGAUAUAUUG---GCAGGCA-------ACAGUGUGCCUGGCAGGCAACCGA----------- ..........((((.(((.((((.(((((((((....)))))...)))).))))....((---.((((((-------......)))))).))))).)))).----------- ( -35.00) >DroEre_CAF1 45433 91 + 1 GUUUGAUAAAUGGUCGCCCAUUAGGCGAGCACUUAAUGGUGCAUAUCGUGUGAUAUAUUG---GCAGGCA-------ACAGUGUGUCUGGCAGGCAACCGA----------- ..........((((.(((.((((.(((((((((....)))))...)))).))))....((---.((((((-------......)))))).))))).)))).----------- ( -32.30) >DroYak_CAF1 46399 102 + 1 GUUUGAUAAAUGGUCGCCCAUUAGGCGAGCACUUAAUGGUGCAUAUCGUGUGAUAUAUUG---GCAGGCA-------ACAGUGUGCCUGGCAGGCAACCGAAGUCCGAAGAC ....(((...((((.(((.((((.(((((((((....)))))...)))).))))....((---.((((((-------......)))))).))))).))))..)))....... ( -35.30) >DroPer_CAF1 48447 101 + 1 AAUUGAUAAAUGGCCCGCUAUUAAAUGAACACUUAAUGGUGCAUAACGUGUGAUAUACUGUCGGCAGGCAGGCAGAGGCAGGGUGGCUGGCACGGAAUCGA----------- ..(((((...(((((.((((((.......((((....))))................(((((.((......))...))))))))))).))).))..)))))----------- ( -26.40) >consensus GUUUGAUAAAUGGUCGCCCAUUAGGCGAGCACUUAAUGGUGCAUAUCGUGUGAUAUAUUG___GCAGGCA_______ACAGUGUGCCUGGCAGGCAACCGA___________ ..........(((..(((.((((.(((((((((....)))))...)))).))))..........((((((.............))))))...)))..)))............ (-18.44 = -18.67 + 0.23)
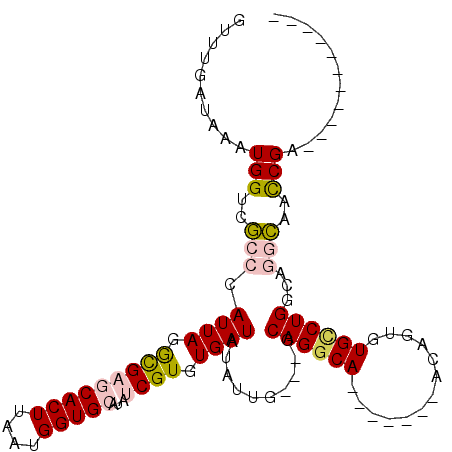
| Location | 15,068,423 – 15,068,514 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 80.99 |
| Mean single sequence MFE | -26.47 |
| Consensus MFE | -16.83 |
| Energy contribution | -17.55 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.872311 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 15068423 91 - 20766785 -----------UCGGAUGCCUGCCAGGCACACUGU-------UGCCUGC---CAAUAUAUCACACGAUAUGCACCAUUAAGUGCUCGCCUAAUGGGCGACCAUUUAUCAAAC -----------..((.(((.((.((((((......-------)))))).---))..(((((....))))))))))..((((((.(((((.....))))).))))))...... ( -30.70) >DroPse_CAF1 48874 101 - 1 -----------UCGAUUCCGUGCCAGACACCCUGCCUCUGCCUGCCUGCCGACAGUAUAUCACACGUUAUGCACCAUUAAGUGUUCAUUUAAUAGCGGGCCAUUUAUCAAUU -----------..(((...(((.((((.........))))...((((((.....(((((........)))))...(((((((....))))))).)))))))))..))).... ( -19.60) >DroSim_CAF1 23136 91 - 1 -----------UCGGUUGCCUGCCAGGCACACUGU-------UGCCUGC---CAAUAUAUCACACGAUAUGCACCAUUAAGUGCUCGCCUAAUGGGCGACCAUUUAUCAAAC -----------..(((.((.((.((((((......-------)))))).---))..(((((....))))))))))..((((((.(((((.....))))).))))))...... ( -30.70) >DroEre_CAF1 45433 91 - 1 -----------UCGGUUGCCUGCCAGACACACUGU-------UGCCUGC---CAAUAUAUCACACGAUAUGCACCAUUAAGUGCUCGCCUAAUGGGCGACCAUUUAUCAAAC -----------..(((.((.((.(((.((......-------)).))).---))..(((((....))))))))))..((((((.(((((.....))))).))))))...... ( -23.60) >DroYak_CAF1 46399 102 - 1 GUCUUCGGACUUCGGUUGCCUGCCAGGCACACUGU-------UGCCUGC---CAAUAUAUCACACGAUAUGCACCAUUAAGUGCUCGCCUAAUGGGCGACCAUUUAUCAAAC (((....)))...(((.((.((.((((((......-------)))))).---))..(((((....))))))))))..((((((.(((((.....))))).))))))...... ( -35.70) >DroPer_CAF1 48447 101 - 1 -----------UCGAUUCCGUGCCAGCCACCCUGCCUCUGCCUGCCUGCCGACAGUAUAUCACACGUUAUGCACCAUUAAGUGUUCAUUUAAUAGCGGGCCAUUUAUCAAUU -----------..(((...(((.(((.....))).........((((((.....(((((........)))))...(((((((....))))))).)))))))))..))).... ( -18.50) >consensus ___________UCGGUUGCCUGCCAGGCACACUGU_______UGCCUGC___CAAUAUAUCACACGAUAUGCACCAUUAAGUGCUCGCCUAAUGGGCGACCAUUUAUCAAAC .............(((((((((..((((............................(((((....)))))((((......))))..))))..)))))))))........... (-16.83 = -17.55 + 0.72)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:24:40 2006