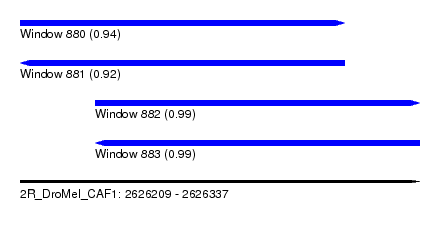
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 2,626,209 – 2,626,337 |
| Length | 128 |
| Max. P | 0.994922 |

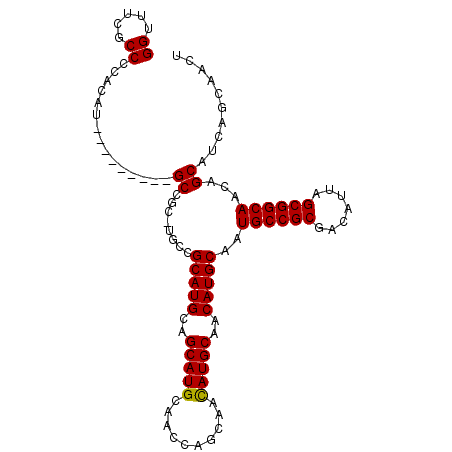
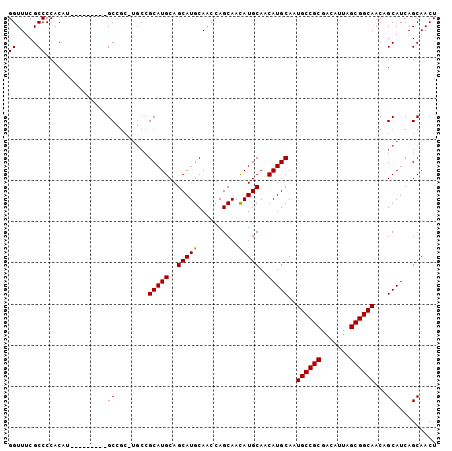
| Location | 2,626,209 – 2,626,313 |
|---|---|
| Length | 104 |
| Sequences | 3 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 88.45 |
| Mean single sequence MFE | -31.07 |
| Consensus MFE | -25.85 |
| Energy contribution | -25.63 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.00 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.943191 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2626209 104 + 20766785 GGUUUCGCCCCACAUGCCCCACAUGCCGCAUGCCGCAUGCAGCAUGCAACCAGCAAUAUGCAACAUGCAAUGCCGCGACAUUAGCGGCAACAGCAUCAGCAACU ((((........((((.....))))..((((((.(....).)))))))))).......(((...((((..((((((.......))))))...))))..)))... ( -33.20) >DroSec_CAF1 60779 88 + 1 GGUUUCGCCCCACAU---------GC-------CGCAUGCAGCAUGCAACCAGCAACAUGCAACAUGCAAUGCCGCGACAUUAGCGGCAACAGCAUCAGCAACU ......((.....((---------((-------.(((((..(((((..........)))))..)))))..((((((.......))))))...))))..)).... ( -28.40) >DroSim_CAF1 58294 95 + 1 GGUUUCGCCCCACAU---------GCCGCUUGCCGCAUGCAGCAUGCAACCAGCAACAUGCAACAUGCAAUGCCGCGACAUUAGCGGCAACAGCAUCAGCAACU (((............---------)))((((((.(((((..(((((..........)))))..)))))..((((((.......))))))...)))..))).... ( -31.60) >consensus GGUUUCGCCCCACAU_________GCCGC_UGCCGCAUGCAGCAUGCAACCAGCAACAUGCAACAUGCAAUGCCGCGACAUUAGCGGCAACAGCAUCAGCAACU ((.....))...............((........(((((..(((((..........)))))..)))))..((((((.......))))))...)).......... (-25.85 = -25.63 + -0.22)

| Location | 2,626,209 – 2,626,313 |
|---|---|
| Length | 104 |
| Sequences | 3 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 88.45 |
| Mean single sequence MFE | -44.03 |
| Consensus MFE | -31.93 |
| Energy contribution | -32.27 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.78 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.915477 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2626209 104 - 20766785 AGUUGCUGAUGCUGUUGCCGCUAAUGUCGCGGCAUUGCAUGUUGCAUAUUGCUGGUUGCAUGCUGCAUGCGGCAUGCGGCAUGUGGGGCAUGUGGGGCGAAACC ..(((((........((((((.......))))))(..((((((.(((((.....(((((((((((....)))))))))))))))).))))))..)))))).... ( -46.80) >DroSec_CAF1 60779 88 - 1 AGUUGCUGAUGCUGUUGCCGCUAAUGUCGCGGCAUUGCAUGUUGCAUGUUGCUGGUUGCAUGCUGCAUGCG-------GC---------AUGUGGGGCGAAACC ..(((((........((((((.......))))))(..((((((((((((.((((....)).)).)))))))-------))---------)))..)))))).... ( -39.00) >DroSim_CAF1 58294 95 - 1 AGUUGCUGAUGCUGUUGCCGCUAAUGUCGCGGCAUUGCAUGUUGCAUGUUGCUGGUUGCAUGCUGCAUGCGGCAAGCGGC---------AUGUGGGGCGAAACC ..(((((.((((...((((((.......))))))..)))).(..(((((((((.((((((((...)))))))).))))))---------)))..)))))).... ( -46.30) >consensus AGUUGCUGAUGCUGUUGCCGCUAAUGUCGCGGCAUUGCAUGUUGCAUGUUGCUGGUUGCAUGCUGCAUGCGGCA_GCGGC_________AUGUGGGGCGAAACC ..(((((..(((...((((((.......))))))..((((((.((((((........)))))).)))))).....................))).))))).... (-31.93 = -32.27 + 0.33)

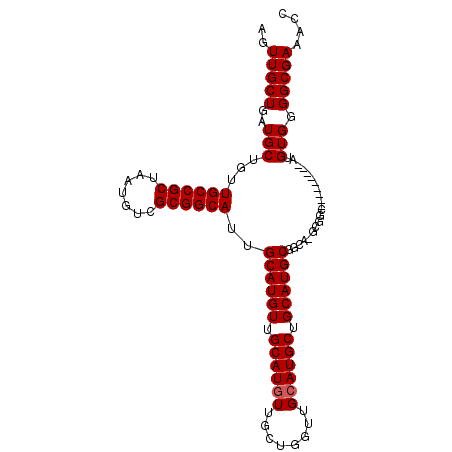
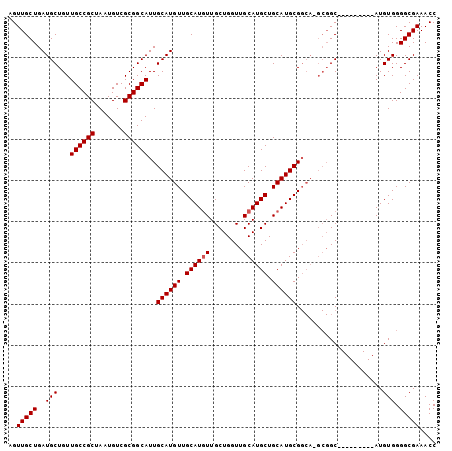
| Location | 2,626,233 – 2,626,337 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 83.25 |
| Mean single sequence MFE | -30.82 |
| Consensus MFE | -21.42 |
| Energy contribution | -21.82 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.34 |
| Structure conservation index | 0.69 |
| SVM decision value | 2.52 |
| SVM RNA-class probability | 0.994922 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2626233 104 + 20766785 GCCGCAUGCCGCAUGCAGCAUGCAACCAGCAAUAUGCAACAUGCAAUGCCGCGACAUUAGCGGCAACAGCAUCAGCAACUUGCUCUAAAAUCUCAACUGCGAGU ((.(.((((.(((((..((((((.....)))...)))..)))))..((((((.......))))))...))))).)).((((((...............)))))) ( -35.36) >DroSec_CAF1 60794 97 + 1 GC-------CGCAUGCAGCAUGCAACCAGCAACAUGCAACAUGCAAUGCCGCGACAUUAGCGGCAACAGCAUCAGCAACUUGCUCCAAAAUCGCAACUGAGAGU ((-------.(((((..(((((..........)))))..)))))..((((((.......))))))...)).((((....((((.........)))))))).... ( -30.40) >DroSim_CAF1 58309 104 + 1 GCCGCUUGCCGCAUGCAGCAUGCAACCAGCAACAUGCAACAUGCAAUGCCGCGACAUUAGCGGCAACAGCAUCAGCAACUUGCUCCAAAAUCGCAACUGCGAGU ...((((((.(((((..(((((..........)))))..)))))..((((((.......))))))...)))..))).((((((...............)))))) ( -35.26) >DroEre_CAF1 59789 104 + 1 GCCACCAGCCACCAGCCACCAGCCACCAGCAACAUGCAACAUGCAAUGCCGCGACAUUAGCGGCAGCAGCAUCAGCAACUUGCUCCCAAAUCGCAACUGCCAGU .......((.....((.....))....(((((..(((...((((..((((((.......))))))...))))..)))..)))))........)).......... ( -25.00) >DroYak_CAF1 58238 85 + 1 -------------------AUGCAACUAGCAACAUGCAACUAGCAAUGCCGCGACAUUAGCGGCAACAGCAUCAGCAACUUGCUCCCAAAUCGCAUCUGCUAGU -------------------.....(((((((..((((.....((..((((((.......))))))...))...(((.....)))........)))).))))))) ( -28.10) >consensus GCC_C__GCCGCAUGCAGCAUGCAACCAGCAACAUGCAACAUGCAAUGCCGCGACAUUAGCGGCAACAGCAUCAGCAACUUGCUCCAAAAUCGCAACUGCGAGU ...........................(((((..(((...((((..((((((.......))))))...))))..)))..))))).................... (-21.42 = -21.82 + 0.40)

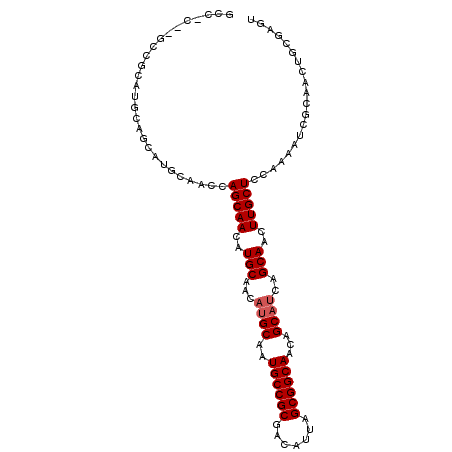
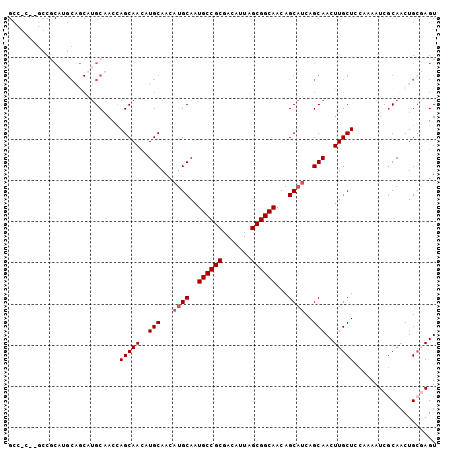
| Location | 2,626,233 – 2,626,337 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 83.25 |
| Mean single sequence MFE | -41.20 |
| Consensus MFE | -28.62 |
| Energy contribution | -30.74 |
| Covariance contribution | 2.12 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.30 |
| Structure conservation index | 0.69 |
| SVM decision value | 2.48 |
| SVM RNA-class probability | 0.994489 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2626233 104 - 20766785 ACUCGCAGUUGAGAUUUUAGAGCAAGUUGCUGAUGCUGUUGCCGCUAAUGUCGCGGCAUUGCAUGUUGCAUAUUGCUGGUUGCAUGCUGCAUGCGGCAUGCGGC .((((....)))).......(((((..(((..((((...((((((.......))))))..))))...)))..))))).(((((((((((....))))))))))) ( -44.10) >DroSec_CAF1 60794 97 - 1 ACUCUCAGUUGCGAUUUUGGAGCAAGUUGCUGAUGCUGUUGCCGCUAAUGUCGCGGCAUUGCAUGUUGCAUGUUGCUGGUUGCAUGCUGCAUGCG-------GC .......((((((((.((((.((((...((....))..))))..)))).)))))))).((((((((.((((((........)))))).)))))))-------). ( -39.20) >DroSim_CAF1 58309 104 - 1 ACUCGCAGUUGCGAUUUUGGAGCAAGUUGCUGAUGCUGUUGCCGCUAAUGUCGCGGCAUUGCAUGUUGCAUGUUGCUGGUUGCAUGCUGCAUGCGGCAAGCGGC ..((((.((((((((.((((.((((...((....))..))))..)))).)))))))).((((((((.((((((........)))))).))))))))...)))). ( -43.90) >DroEre_CAF1 59789 104 - 1 ACUGGCAGUUGCGAUUUGGGAGCAAGUUGCUGAUGCUGCUGCCGCUAAUGUCGCGGCAUUGCAUGUUGCAUGUUGCUGGUGGCUGGUGGCUGGUGGCUGGUGGC ...((((((.((((((((....))))))))....))))))(((((((.....(((((((...)))))))..((..(..((.(....).))..)..))))))))) ( -45.30) >DroYak_CAF1 58238 85 - 1 ACUAGCAGAUGCGAUUUGGGAGCAAGUUGCUGAUGCUGUUGCCGCUAAUGUCGCGGCAUUGCUAGUUGCAUGUUGCUAGUUGCAU------------------- ((((((((((((((((....(((((...((....))...((((((.......))))))))))))))))))).)))))))).....------------------- ( -33.50) >consensus ACUCGCAGUUGCGAUUUUGGAGCAAGUUGCUGAUGCUGUUGCCGCUAAUGUCGCGGCAUUGCAUGUUGCAUGUUGCUGGUUGCAUGCUGCAUGCGGC__G_GGC ....((((((((((((....(((((..(((..((((...((((((.......))))))..))))...)))..)))))))))))).))))).............. (-28.62 = -30.74 + 2.12)


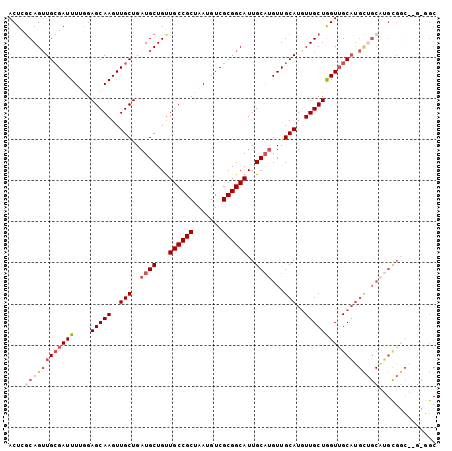
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:39:12 2006