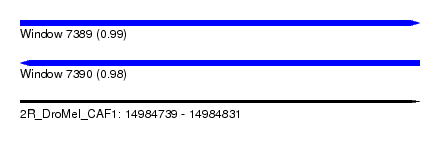
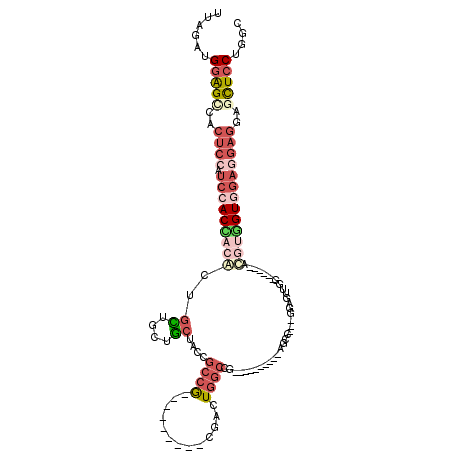
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,984,739 – 14,984,831 |
| Length | 92 |
| Max. P | 0.987324 |

| Location | 14,984,739 – 14,984,831 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 67.54 |
| Mean single sequence MFE | -41.26 |
| Consensus MFE | -19.88 |
| Energy contribution | -21.60 |
| Covariance contribution | 1.72 |
| Combinations/Pair | 1.42 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.48 |
| SVM decision value | 2.07 |
| SVM RNA-class probability | 0.987324 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14984739 92 + 20766785 GCCAGGAGCUCCUCCUCCACCACGU------CCACCUCC---GGCU----------CGGCCAGUCG---------UGGCGGUAGCAGCAGCAGCGUGGUGGAUGGAGUGGCUCCAUCUAA ....(((((..((((((((((((((------....((.(---.(((----------(.((((....---------)))).).))).).))..)))))))))).))))..)))))...... ( -49.10) >DroSec_CAF1 2711 92 + 1 GCCAGGAGCUCCUCCUCCACCACGU------CCACCUCC---CGCU----------CGGCCAGUCG---------CGGCGGUAGCAGUAGCAGCGUGGUGGAUGGAGUGGCUCCAUCUAA ....(((((..((((((((((((((------.......(---((((----------(((....)))---------.)))))..((....)).)))))))))).))))..)))))...... ( -49.40) >DroWil_CAF1 12069 114 + 1 ---AGGAAGUCGUUCGUCACAUCAUUCGACUCCAACACCAGCUGCU---CGAACAACAUCCAGUCGAGCUGUAAGCGGCGGUAGUAGUAAUAGUGUUGUCGAUGGGGUUACCACAUCCCG ---.(((.((((...(......)...))))))).......((.(((---(((.(........))))))).))...((((..((.((....)).))..))))..(((((......))))). ( -32.20) >DroYak_CAF1 2722 92 + 1 GCCAGGAGCUCCUCGUCCACCAUGU------CCAACUCC---GCCU----------CCGCCAGUCG---------UGGCGGUAGCAGCAGCAGUGUAGUGGAUGGAGUGACUCCAUCUAA ....(((((.(..(((((((.....------........---((..----------((((((....---------))))))..)).(((....))).)))))))..).).))))...... ( -34.70) >DroAna_CAF1 14007 92 + 1 GCCCGGAGUUCCUCGUCGAC---GU------CGAACUCC---ACAUCCG-------CGGGAAGUCG---------CGGCGGAAGCAGCAACAGUGUCGUGGACGGAGUGGCUCCGUCCAA ((..(((((((..((....)---).------.)))))))---.....((-------(((....)))---------))((....)).))..........(((((((((...))))))))). ( -40.90) >consensus GCCAGGAGCUCCUCCUCCACCACGU______CCAACUCC___GGCU__________CGGCCAGUCG_________CGGCGGUAGCAGCAGCAGUGUGGUGGAUGGAGUGGCUCCAUCUAA ....((((((.((((((((((((((.....................................((((.........))))....((....)).)))))))))).)))).))))))...... (-19.88 = -21.60 + 1.72)

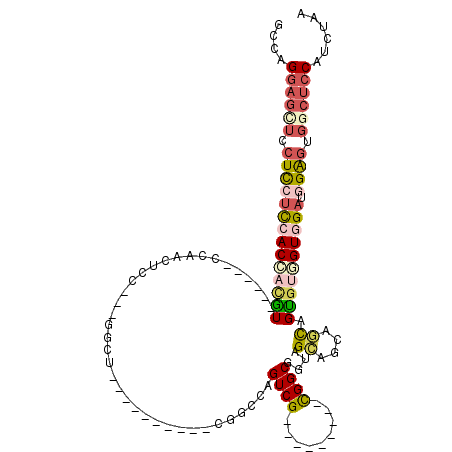
| Location | 14,984,739 – 14,984,831 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 67.54 |
| Mean single sequence MFE | -43.52 |
| Consensus MFE | -15.48 |
| Energy contribution | -18.56 |
| Covariance contribution | 3.08 |
| Combinations/Pair | 1.38 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.36 |
| SVM decision value | 1.97 |
| SVM RNA-class probability | 0.984335 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14984739 92 - 20766785 UUAGAUGGAGCCACUCCAUCCACCACGCUGCUGCUGCUACCGCCA---------CGACUGGCCG----------AGCC---GGAGGUGG------ACGUGGUGGAGGAGGAGCUCCUGGC ......(((((..((((.((((((((((..((.(((((.(.((((---------....)))).)----------)).)---)).))..)------.)))))))))))))..))))).... ( -51.00) >DroSec_CAF1 2711 92 - 1 UUAGAUGGAGCCACUCCAUCCACCACGCUGCUACUGCUACCGCCG---------CGACUGGCCG----------AGCG---GGAGGUGG------ACGUGGUGGAGGAGGAGCUCCUGGC ......(((((..((((.((((((((((..((.(((((.(.((((---------....)))).)----------))))---)..))..)------.)))))))))))))..))))).... ( -53.40) >DroWil_CAF1 12069 114 - 1 CGGGAUGUGGUAACCCCAUCGACAACACUAUUACUACUACCGCCGCUUACAGCUCGACUGGAUGUUGUUCG---AGCAGCUGGUGUUGGAGUCGAAUGAUGUGACGAACGACUUCCU--- .((((((.((....))))))..((((((((...........((.(((((((((((.....)).)))))..)---))).))))))))))((((((..((......))..)))))))).--- ( -31.10) >DroYak_CAF1 2722 92 - 1 UUAGAUGGAGUCACUCCAUCCACUACACUGCUGCUGCUACCGCCA---------CGACUGGCGG----------AGGC---GGAGUUGG------ACAUGGUGGACGAGGAGCUCCUGGC ......(((((..(((..(((((((..(.(((.(((((.((((((---------....))))))----------.)))---))))).).------...))))))).)))..))))).... ( -42.90) >DroAna_CAF1 14007 92 - 1 UUGGACGGAGCCACUCCGUCCACGACACUGUUGCUGCUUCCGCCG---------CGACUUCCCG-------CGGAUGU---GGAGUUCG------AC---GUCGACGAGGAACUCCGGGC .(((((((((...)))))))))((((...((((..((((((((((---------((......))-------))).)).---))))).))------))---))))....((....)).... ( -39.20) >consensus UUAGAUGGAGCCACUCCAUCCACCACACUGCUGCUGCUACCGCCG_________CGACUGGCCG__________AGCC___GGAGUUGG______ACGUGGUGGAGGAGGAGCUCCUGGC ......(((((..((((.(((((((((..((....))....((((.............))))..................................)))))))))))))..))))).... (-15.48 = -18.56 + 3.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:24:22 2006